ORIGINAL RESEARCH article
Identification of apple leaf diseases by improved deep convolutional neural networks with an attention mechanism.

- 1 College of Mechanical and Electronic Engineering, Northwest A&F University, Xianyang, China
- 2 Key Laboratory of Agricultural Internet of Things, Ministry of Agriculture and Rural Affairs, Xianyang, China
- 3 Shaanxi Key Laboratory of Agricultural Information Perception and Intelligent Services, Xianyang, China
- 4 College of Information Engineering, Northwest A&F University, Xianyang, China
The accurate identification of apple leaf diseases is of great significance for controlling the spread of diseases and ensuring the healthy and stable development of the apple industry. In order to improve detection accuracy and efficiency, a deep learning model, which is called the Coordination Attention EfficientNet (CA-ENet), is proposed to identify different apple diseases. First, a coordinate attention block is integrated into the EfficientNet-B4 network, which embedded the spatial location information of the feature by channel attention to ensure that the model can learn both the channel and spatial location information of important features. Then, a depth-wise separable convolution is applied to the convolution module to reduce the number of parameters, and the h-swish activation function is introduced to achieve the fast and easy to quantify the process. Afterward, 5,170 images are collected in the field environment at the apple planting base of the Northwest A&F University, while 3,000 images are acquired from the PlantVillage public data set. Also, image augmentation techniques are used to generate an Apple Leaf Disease Identification Data set (ALDID), which contains 81,700 images. The experimental results show that the accuracy of the CA-ENet is 98.92% on the ALDID, and the average F1-score reaches .988, which is better than those of common models such as the ResNet-152, DenseNet-264, and ResNeXt-101. The generated test dataset is used to test the anti-interference ability of the model. The results show that the proposed method can achieve competitive performance on the apple disease identification task.

Introduction
The apple industry is one of the most important fruit industries in China. However, the frequent occurrence of apple leaf diseases may seriously restrict the healthy and stable development of the apple industry. At present, the diseases of a large number of industrialized apple orchards mainly rely on human vision for recognition, which requires a high degree of reliance on disease experts. The identification task is huge, especially since the visual inspection of fruit farmers or experts is prone to misjudgment due to their subjective perception and visual fatigue, and it is difficult to meet the demand for high-precision identification for intelligent orchards ( Dutot et al., 2013 ). The problems previously discussed will lead to a large lag in the tracking management process of orchard diseases, which causes the improper use of pesticides and reduces the quality of fruit. Therefore, the accurate identification of diseases is of great significance to improve the yield and quality of apples and to cultivate disease-resistant varieties.
With the development of computer vision, machine learning techniques have been widely used in the agricultural field in recent years, and a series of approaches have been achieved in crop disease identification ( Aravind et al., 2018 ; Kour and Arora, 2019 ; Mohammadpoor et al., 2020 ). In recent years, the main techniques, which are widely used in crop disease identification include artificial neural network (ANN) ( Sheikhan et al., 2012 ), the K Nearest Neighbors (KNN) algorithm ( Guettari et al., 2016 ), random forests (RF) ( Kodovsky et al., 2012 ), and so on. For example, Wang et al. (2019) proposed a method for identifying cucumber powdery mildew based on a visible spectrum by extracting the spectral features and training a Support Vector Machine (SVM) classifier to establish a classification model, optimizing the radial basis kernel function, and the recognition accuracy of the method reached 98.13%. In contrast, Prasad et al. (2016) proposed a mobile client-server architecture for leaf disease detection and diagnosis based on the combination of a Gabor Wavelet Transform (GWT) and a Gray-Level Co-occurrence Matrix (GLCM). The mobile terminal captures the object image and then transmits it to the server after pre-processing. The server then performs GWT-GLCM feature extraction and classification based on the KNN algorithm. The system can monitor farmland information through the mobile terminal at any stage. Although the previously discussed studies achieved outstanding performances in disease identification tasks, the low-level feature representations extracted from them are limited to intuitive shallow features, such as the colors, textures, and shapes of the images. Thus, it is difficult to achieve competitive performance on apple leaf disease identification tasks.
Compared with machine learning algorithms that require cumbersome image pre-processing and feature extraction ( Kulin et al., 2018 ; Zhang et al., 2018b ), convolutional neural networks (CNNs) can directly learn robust high-level feature representations of apple diseases from images. The extracted high-level feature representation is richer and better compared with the method of manually extracting features; therefore, CNNs have achieved excellent results in multiple visual tasks ( Ren et al., 2017 ; Liu et al., 2018 ; Bi et al., 2020 ). In recent years, with the continuous emergence of advanced deep learning architectures such as the ResNet ( He et al., 2016 ), ResNeXt ( Xie et al., 2017 ), and DenseNet ( Huang et al., 2017 ), the recognition accuracy and speed are constantly being refreshed on the public dataset, ImageNet. In order to solve the problem of the mobile deployment of the model, scholars have proposed various lightweight architectures, such as Xception ( Chollet, 2017 ), MobileNet ( Howard et al., 2017 ; Sandler et al., 2018 ), ShuffleNet ( Ma et al., 2018 ; Zhang et al., 2018a ), and so on. In order to provide a stable, efficient, low-cost, and highly intelligent disease identification method, Chao et al. (2020) proposed that the XDNet combined with DenseNet and Xception can enhance the feature extraction capability of the model. The model achieved an accuracy of 98.82% in identifying five apple leaf diseases with fewer parameters. Liu et al. (2020) adopted the Inception structure and introduced a dense connection strategy to build a new neural network model, which realized the real-time and accurate identification of six different kinds of grape leaf diseases. In addition, Ramcharan et al. (2019) deployed a trained cassava disease recognition model for a mobile terminal. Tests under natural conditions in the field found that complex conditions, such as different angles, brightness, and the occlusion of the image taken, could adversely affect the performance of the model, which also proves that image classification under the complex background of the field is challenging.
An attention mechanism can provide a novel solution for feature extraction. The attention mechanism can assign larger weights to regions of interest and smaller weights to backgrounds and extract information that contributes more to classification to optimize the model and to make judgments that are more accurate. In other studies, attention mechanisms have achieved excellent performance in tasks, such as classification, detection, and segmentation ( Hu et al., 2018 ; Karthik et al., 2020 ; Mi et al., 2020 ; Hou et al., 2021 ). Inspired by the above researches, this study proposes a new CNN for apple diseases recognition. The main contributions and innovations of this study are summarized as follows:
1. A new Apple Leaf Disease Identification Data set (ALDID) is generated by using image generation techniques. In order to enhance the generalization performance of the model, image augmentation techniques are used to expand the data set and simulate apple leaf disease images collected under different conditions, laying a foundation for the training of the model.
2. A novel attention-based apple leaf disease recognition model, namely, the Coordination Attention EfficientNet (CA-ENet), is proposed. A network search technique is first used to determine the optimal structure of the model, and the optimal parameters of network depth, width, and input image resolution are obtained. Then, the deep separable convolution is applied to the coordination attention convolution (CA-Conv) infrastructure to greatly reduce the number of parameters and avoid an overfitting problem. Finally, a coordinated attention block is embedded in the infrastructure to realize the integration of characteristic channel information and spatial information attention and to strengthen the learning ability of the model for important information in the lesion area.
The remainder of the study is organized as follows: In section Materials and Methods, the detailed information of the dataset is introduced and expanded by data augmentation techniques. The model proposed in this study and the related content of attention visualization is introduced in detail. The section Results and Discussion presents the experiments for evaluating the performance of the model and analyzes the results of the experiments, discussed the impact of data augmentation and external interference on the performance of the model. The last section, Conclusion and Future Work, summarizes the work of this study and prospects for further research.
Materials and Methods
This section introduces the materials and methods used in the study in detail, including the collected apple diseased leaf images and the ALDID established after augmentation. It also presents the proposed model and the attention visualization method.
Image Acquisition
The study was conducted from July 2020 to October 2020, at the apple planting experimental station of the Northwest A&F University in Qianxian County, Shaanxi province. By using a variety of different types of mobile devices, a huge number of field environment apple leaf images under different angles and distances are collected. There are a total of 5,170 disease images with a resolution of 3,000 × 3,000 pixels, including those of five species of the Glomerella leaf spot ( Colletotrichum fructicola ), Apple leaf mites ( Panonychus ulmi ), Mosaic (Apple mosaic virus), Apple litura moth ( Spodoptera litura Fabricius ), and Healthy leaves. In addition, 3,000 disease images under a single background of three kinds of laboratories, namely, Black rot ( Physalospora obtuse ), Scab ( Venturia inaequalis ), and Rust ( Gymnosporangium yamadai ), were collected from the public dataset PlantVillage. The above two data sets are shuffled and mixed to generate the original data set of common apple diseases.
Figure 1 shows random samples of each category in the data set. There are a large number of complex background images in the data set. At the same time, it can be seen that Apple litura moth (G) and Apple leaf mites (H) leaves have relatively similar geometric features. The difference between the two diseases can be expressed as a fine-grained image classification problem. A variety of different forms of samples can increase the diversity of the data set, making it closer to various different situations that may occur in the real situation. However, it also constitutes a greater test for the image classification task and puts forward higher requirements for the comprehensive performance of the model.
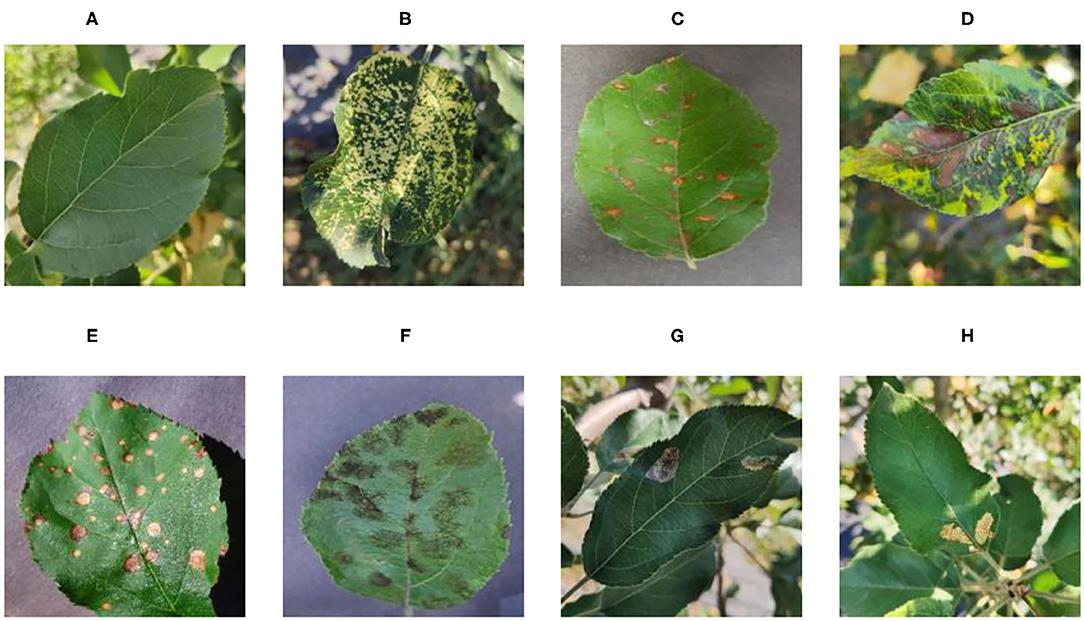
Figure 1 . Eight common apple leaf disease types. (A) Healthy, (B) Mosaic, (C) Rust, (D) Glomerella leaf spot, (E) Black rot, (F) Scab, (G) Apple litura moth, and (H) Apple leaf mites.
Image Augmentation
When acquiring the apple disease images, the samples obtained varied in the apple leaf growth position, weather condition, shooting angle, and there are interference factors such as equipment noise. In order to enable the model to learn as many irrelevant patterns as possible and avoid overfitting problems, the images of the dataset need to be expanded and normalized.
In the data expansion, Gaussian blurring, contrast enhancement by 30% and decrease by 30%, and brightness enhancement by 30% and decrease by 30% are adopted to simulate different weather conditions for all samples of the original dataset. The images are also rotated by 90°, 270°, a horizontal flip, and a vertical flip to simulate the change of shooting angle, then the original data set is added. A Mosaic disease image is randomly selected to enhance and display the effect as shown in Figure 2 . Table 1 represents the structure information of the ALDID. It can be seen from Table 1 that the sample distribution is balanced after image expansion, which is in line with the actual application scenario. It can ensure that the model extracts different features of each category in a balanced manner, ensuring its correct training and avoiding overfitting. This study also divides the ALDID according to the ratio of training set: validation set = 4:1 for model training and validation. The training set is used to train the model, and the validation set is used to check whether the model training process converges normally and whether there is an overfitting problem.

Figure 2 . Image enhancement example of the mosaic disease. (A) Original image, (B) Gaussian blur, (C) 90° rotation, (D) High contrast, (E) Low contrast, (F) 270° rotation, (G) Horizontal symmetry, (H) Vertical symmetry, (I) High brightness, and (J) Low brightness.
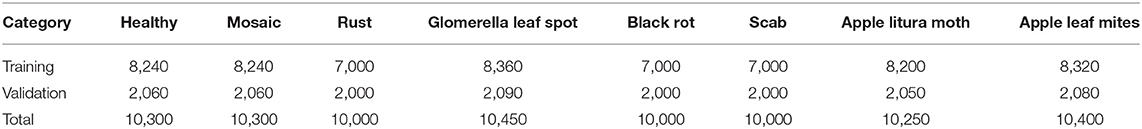
Table 1 . The composition of apple leaf disease identification data set (ALDID).
During the training process, a large fluctuation of the feature value range will affect the convergence of the model, which is not conducive to the model learning different feature differences, and the images need normalization. In order to test the stability of the model, 500 images were randomly selected from each type of disease image in the original data set, and a total of 4,000 images were selected from eight different diseases. After scrambling these 4,000 images, five different interference factors, namely, Gaussian noise, salt and pepper noise, 180° rotation, 30% sharpness enhancement, and 30% sharpness reduction were randomly added, and a Model Robustness Test Data set (MRTD) was generated. After the training process is completed, the MRTD is then used to test the model to verify the effect of the model training. The above work laid the foundation for the use of the model.
CA-ENet Network
The existing CNN methods of increasing network depth, width, and input image resolution can obtain richer and higher fine-grained features, but, there will be serious problems such as gradient disappearance and model degradation. The problem is that only changing a single variable cannot achieve better results. The basic network architecture EfficientNet-B0 ( Tan and Quoc, 2019 ), which uses neural architecture search (NAS) techniques to optimize the above three factors at the same time, balances the three dimensions of depth, width, and resolution, and can be further adjusted by the scaling factor. Therefore, in this study, we use the EfficientNet architecture as the feature extraction network.
Different types of apple leaf diseases have different morphological characteristics with regard to lesions, but there is a high degree of similarity between certain types of diseases, which means apple disease classification can be viewed as a fine-grained image classification problem, and existing models still have difficulty achieving satisfactory results. Therefore, in order to enhance model effectiveness, attention to the lesion area is the key to solving this problem. The widely used channel attention mechanism, SENet ( Hu et al., 2018 ), has a significant effect on improving final performance, but this operation ignored the location information of the features, which is also important for generating spatial selective attention maps. In order to identify these differences, the CA-ENet is proposed to achieve real-time and accurate apple disease identification. The overall structure of the model is shown in Figure 3 .
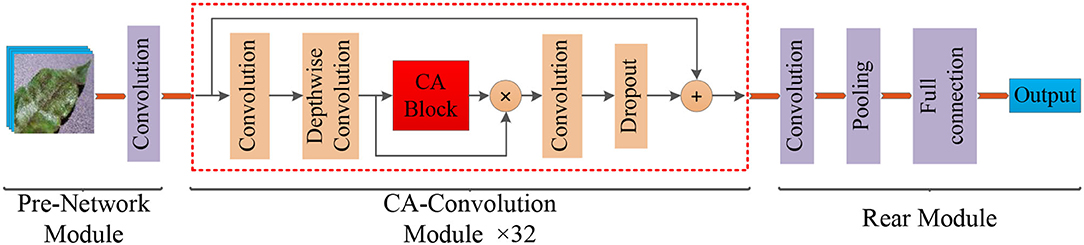
Figure 3 . Structure of the Coordination Attention EfficientNet (CA-ENet) for apple disease identification.
The model mainly included three parts: the pre-network for the Batch Normalization of input images, the backbone network CA-Conv for feature extraction, and the rear part that outputs the recognition result through the fully connected layer. Pre-network uses a layer of 3 × 3 ordinary convolutions with a step of 1 to perform the convolution operation on the input image, the input image resolution is 380 × 380, and the feature map with the depth of the output feature matrix of 48 is obtained. Then, the obtained feature matrix are input into the 32 CA-Conv module embedded with the CA block. Finally, the 3 × 3 ordinary convolutions and pooling are used to further abstract features and then output through a fully connected layer with eight nodes.
During the model optimization process, a NAS technique is used to search for the optimal model structure. The operation process can be abstractly expressed as Equation (1):
where ⊙ is the multiplication symbol. F i L i means arithmetic operation, it is repeatedly executed L i times in the operation F i . X is the input feature matrix. ( H i , W i , and C i ) represents the height, width, and output channels of X . The NAS process can be optimized by adding the constraints of model accuracy, parameter, and calculation amount with Equations (2) and (5).
The d, w , and r are the sparseness that scales the depth, width, and resolution of the network, respectively, the tar_memory and tar_flops are the constraints on the number of parameters and calculations. Through the above optimization calculation, the best d, w , and r values of the EfficientNet-B0 structure can be obtained, and on this basis, the magnification factors d and w of EfficientNet-B4 are 1.8 and 1.4, respectively, and the input image resolution r is 380 × 380 pixels. From the discussed method, the optimal CA-ENet structure parameters can be calculated and are shown in Table 2 .
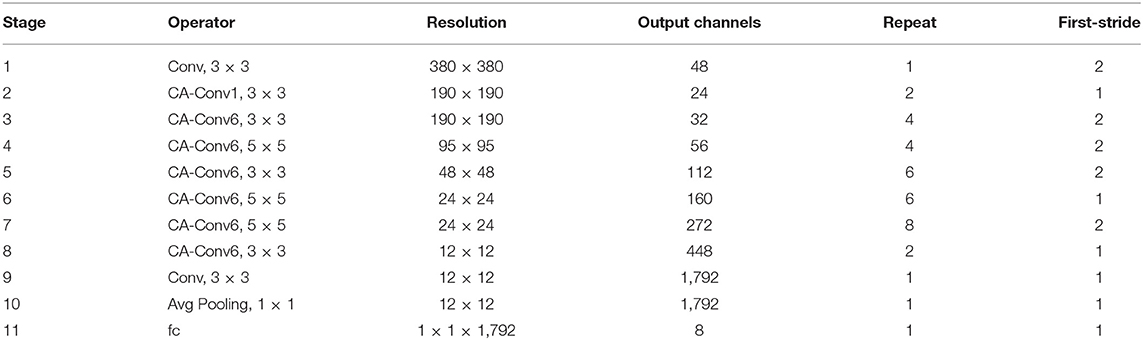
Table 2 . Details about coordination attention EfficientNet (CA-ENet).
The operators in Table 2 perform arithmetic operations on the input features. The magnification of each CA-Conv6 in Stage 3–Stage 8 is 6; that is, in the first layer of convolution, the depth of the feature matrix of the input layer is increased to 6 times of the input, and the size of the convolution kernel is 3 × 3 or 5 × 5. The resolution, output channels, and repeat correspond to the resolution of the input layer, the depth of the output feature matrix, and the number of repetitions of the layer structure in the depth direction. The steps given by first-stride are only for the first layer structure of each stage, and the steps of the other layer structures are all 1. The network is composed of seven-stage CA-Conv blocks, and its structure is shown in Figure 4 . First, the input feature matrix is sent to CA-Conv through an ordinary 1 × 1 convolution for dimension upgrade. After the h-swish activation function, the feature is extracted through the deep separable convolution with a convolution kernel size of k × k (k = 3 or 5) and a step of 1 or 2. The use of a deep separable convolution structure greatly reduces the number of model parameters, and at the same time, can play an important role in avoiding model overfitting. Then, the obtained feature matrix is divided into two branches, one of which is assigned a weight to each channel by a Coordinate Attention Block (CAB), and another one without any processing is multiplied by the two weights passed through the CAB to obtain the weighted feature matrix. Finally, the dimension is reduced by 1 × 1 convolution and output to the subsequent structure after adding with the input feature matrix.
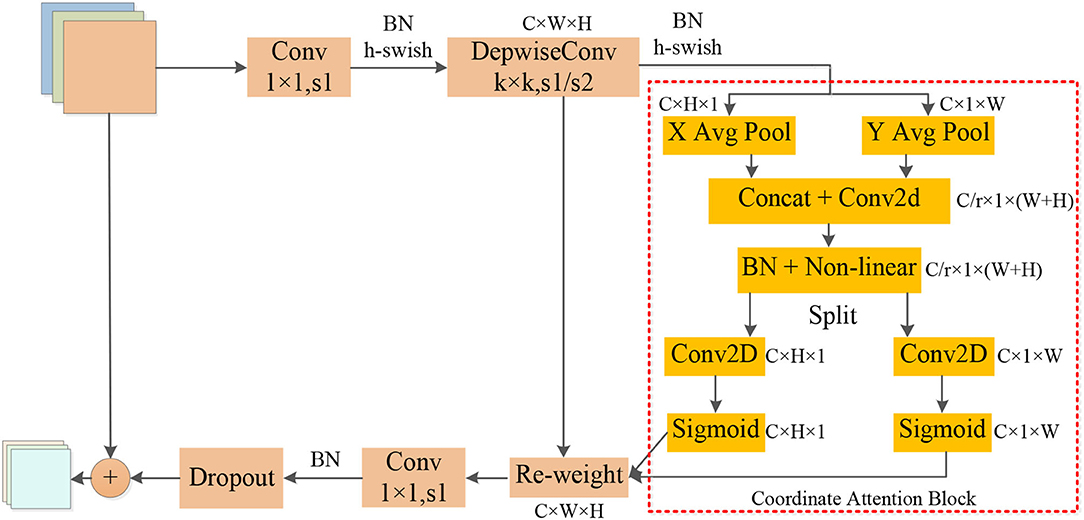
Figure 4 . Structure of coordination attention convolutional (CA-Conv).
The global pooling method can compress the global spatial information into the channel descriptor, but this results in a lack of location information. In order to capture the precise location information of the features, in the CAB in Figure 4 , the global pooling is decomposed into two one-dimensional feature encoding processes according to Equation (6). Furthermore, two one-dimensional average pooling operations along the horizontal and vertical directions are used to aggregate the input features into two separate direction-aware feature maps. This operation captures both direction-aware and position-sensitive information, thus enabling the model to locate the region of interest more accurately. The generated two separate direction-aware feature maps are concatenated in the depth direction, and the feature channel attention weight is generated through a 1 × 1 convolution compression channel, and the position information is embedded in the channel attention. Then, the Batch Nomalization (BN) operation is applied to the feature matrix and divided into two parts through a non-linear activation function, the feature depth is adjusted to be consistent with the input feature through 1 × 1 convolution, and the position information is saved in the generated attention map. Finally, the weights of the two attention maps are multiplied by the input features to strengthen the feature representation of the attention region and improve the ability of the network to locate the regions of interest accurately.
As the above-mentioned information embedding method can directly obtain the global receptive field and encode the accurate position information, so the transformation operation is performed on it using the 1 × 1 convolution transformation function F 1 . As shown in Equation (7), [ z h , z w ] is the splicing operation along a spatial dimension, δ is the non-linear activation function, and f is the intermediate feature map that encodes the spatial information in both horizontal and vertical directions. Then, through two 1 × 1 convolutions, f h and f w are transformed into tensors with the same number of channels, respectively. As shown in Equations (8) and (9), attention weights can be calculated, and the output of the CA block after the Re-weight is calculated by Equation (10).
In order to reduce the amount of calculation and speed up reasoning while ensuring the effect of the activation function, a new activation function, h-swish, is applied into CA-Conv ( Howard et al., 2019 ). The activation functions of sigmoid and h-sigmoid are shown in Equations (11) and (12). It can be seen from Figure 5 that the above two activation functions are relatively close and the calculation process of h-sigmoid is more concise, so h-sigmoid can be used to replace sigmoid in Equations (13) and (14). Figure 6 shows the approximation of the effect of h-swish on the swish activation function. It can be seen that the two curves are basically the same, and the calculation speed of the h-swish is faster.
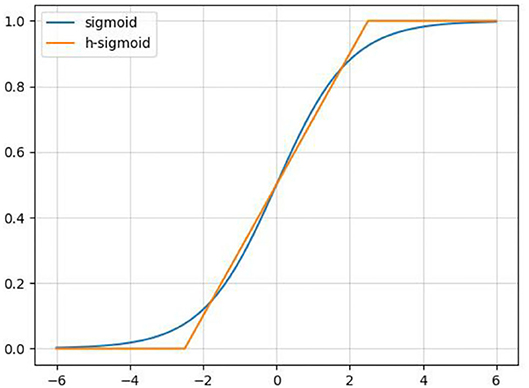
Figure 5 . Schematic diagram of the sigmoid and h-sigmoid activation functions.
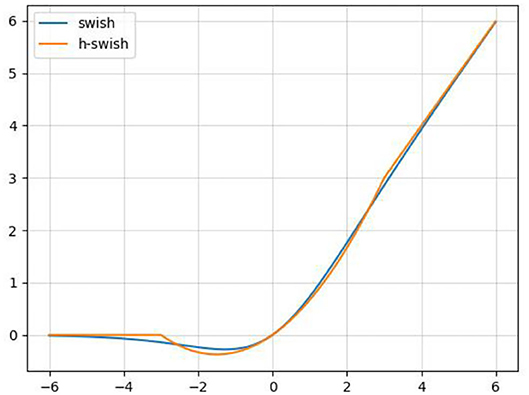
Figure 6 . Schematic diagram of the swish and h-swish activation functions.
Experimental Results and Discussion
Model training details.
In order to verify the performance of the proposed method, a proposed network is trained via the ALDID. Thus, the proposed method is realized on the Pytorch 1.7.1 deep learning framework, while all experiments were conducted on an Intel ® Xeon(R) Gold 5217 CPU@3.00 GHz server equipped with an NVIDIA Tesla V100 (32GB) GPU. The operating system is Ubuntu 18.04.5 LTS 64. In order to accelerate the model convergence while keeping stable training, the initial learning rate is set to .01, and it decays according to the cosine learning rate change curve during the training process, and finally decays to .001. The number of training iterations for all models is 50 epochs.
Performance of Proposed CA-ENet
In order to evaluate the performance of the proposed method, multiple state-of-the-art methods were applied to the MRTD. In order to ensure that the results are comparable, the same training strategy was used. The test result is visually displayed with a confusion matrix. In order to facilitate the display of labels, the full names of some diseases are abbreviated. In this case, “GLS” in the confusion matrix stands for Glomerella leaf spot, “ALM” stands for Apple litura moth, and “ALMS” stands for Apple leaf mites.
Figure 7 can intuitively show the classification performance of the Coordination Attention EfficientNet, with the final accuracy reaching 98.92%. The misclassification mainly occurred between Apple leaf mites and Apple litura moth and between Apple litura moth and Healthy leaves. The main feature of the apple leaf mites is that the damaged leaves show many dense chlorosis gray-white spots. In contrast, after being damaged by apple litura moth, the insect spots formed on the leaves were elliptical and dense, and the leaf surface was wrinkled. The above two kinds of leaf spots have certain similarities in geometric and color characteristics, leading to misjudgment. Furthermore, affected by the complex background, a small number of leaves damaged by apple litura moths were mistakenly identified as healthy leaves. It can be seen that accurate recognition in a complex background has been a great challenge, but the number of misjudgments in this model is still within an acceptable range and can be maintained at a low level. The proposed CA-Conv structure can extract richer fine-grained features of the image and perceive the regions of interest with a higher degree of attention. It can also be seen that the model shows a good recognition effect and has strong robustness to the problem of apple leaf disease recognition.
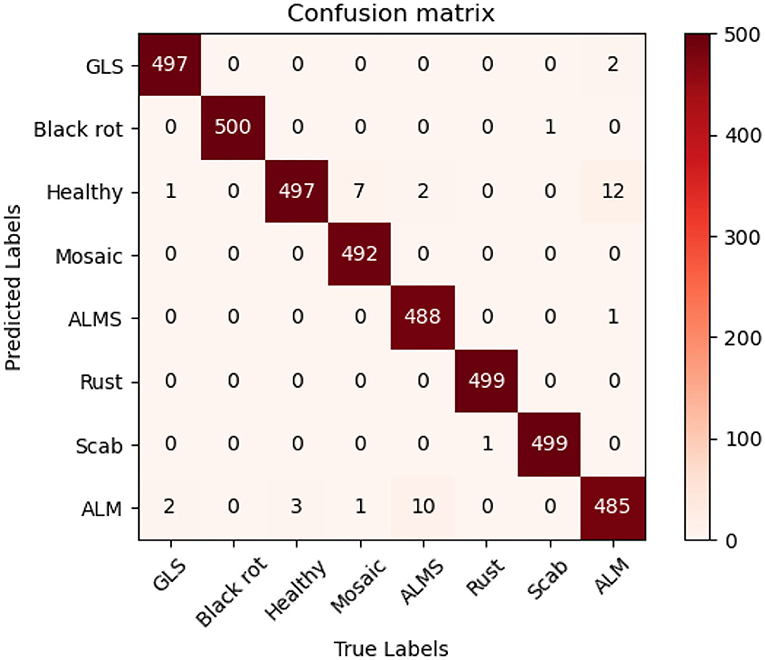
Figure 7 . Confusion matrix of the CA-ENet.
Performance Comparison
The performance comparison between CA-ENet and the standard method is shown in Table 3 . It can be seen from Table 3 that the proposed model has the best recognition performance on MRTD, with an accuracy of 98.92%. In this study, multiple metrics including accuracy, precision, recall, F1-score, parameter, and calculation are used as evaluation indicators. ResNet-152 takes advantage of the residual structure to make sure it has a strong feature learning ability, so it can reach an accuracy of 93.75%. The Dense Block, the basic structure of DenseNet-264, also has the advantages of enhanced feature propagation and incentive feature reuse, making it achieve a higher accuracy rate with nearly half of the parameters of ResNet-152. Furthermore, the accuracy of ResNeXt-101 reaches 95.67%, which is due to the use of grouped convolution, so it can achieve better results with fewer convolutional layers than ResNet-152. Although this structure can improve the final accuracy, the degree of network fragmentation is very high due to the existence of a large number of parallel branches, which greatly reduces the computation efficiency of the model.
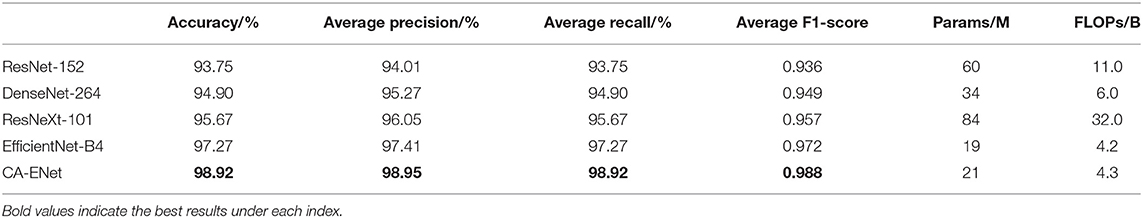
Table 3 . Performance comparison of the CA-ENet with other classical networks.
EfficientNet uses NAS techniques to simultaneously search and optimize model depth, width, and input image resolution, and rationally expand the model architecture to achieve a high degree of coordination of structural proportions. It has obvious advantages in extracting more robust and reliable feature representations and can reach an overall accuracy of 97.27%. The strong learning ability of the CA module in CA-Conv may cause attention drift and affect model convergence, while the inverted residual structure in CA-Conv can suppress features that are not conducive to classification, ensuring model stability while further improving the recognition performance, and the effectiveness of the attention mechanism is verified.
Traditional CNNs do not distinguish the importance of information when extracting disease features, and there is a large number of convolutions that repeatedly extract low-contribution information, which causes a waste of computation resources. The attention mechanism can automatically extract high-contribution feature components, with only small parameters and calculations increases. The experimental results also show that, in the identification of apple leaf diseases, the proposed CA-ENet model is superior to other models in all evaluation indicators with fewer parameters and can classify apple disease images more accurately.
Effect of Data Augmentation on Identification Performance for Each Class
A variety of data expansion methods is used in the ALDID to improve the anti-interference ability of the model in complex situations and prevent the problem of overfitting. In order to verify the effect of data augmentation, a set of comparative experiments is designed to evaluate its impact on the final classification performance. Table 4 shows the accuracy, recall, and F1-score performance indicators of the proposed model for each category on the MRTD. The first row of values in each performance index is the performance obtained after training on the original dataset, and the second row of values is the performance obtained after training on the ALDID. It can be seen from Table 4 that the image diversity of the original data set is insufficient, and the average F1-score of the proposed method on the original dataset is 0.969, which is slightly lower than the performance of the model obtained on the ALDID, but it can still accurately classify apple leaf diseases. The results show that the augmented data set is closer to the actual situation, the ability of the model to adapt to complex scenes is enhanced, and the anti-interference ability is improved to a certain extent. The leverage of the deep separable convolution can effectively reduce the number of model parameters and greatly increase training speed.
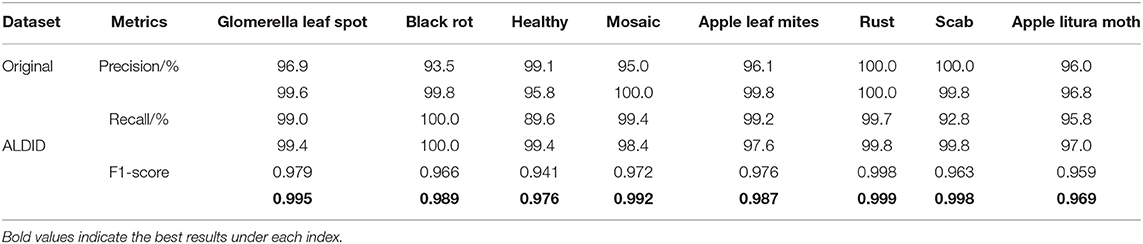
Table 4 . Performance of the CA-ENet before and after data augmentation.
Feature and Network Attention Visualization
Understanding and analyzing the hidden layer structure of the model is an important method to comprehensively recognize the proposed network structure. CNNs are usually trained in the form of black-box testing and the evaluations of model performance are limited to the final accuracy and other indicators, which have certain deficiencies. Visualization techniques are the way to explore how CNNs learn features and distinguish categories. So, this section uses the visualization of layer activation and class activation heatmaps to analyze the performance of the proposed model. The visualization of layer activation helps to understand how the continuous convolutional layer performs feature extraction and completes the conversion of input features. Figures 8 , 9 show the output features of the first 20 channels of the CA-Conv structure in the first and last layers of the model, respectively. The given example category is apple leaf mites. In the superficial features of the model, it is obvious that the lesion area and the background are separated, and the characteristics of the disease location can be accurately extracted. The model has high efficiency in extracting deep features and only contains a few failed convolutions. The channel output features given here are all valid. Therefore, the stacking of the CA-Conv structure does not affect feature learning ability of the model, and the adopted separable convolution can effectively reduce the feature redundancy and lead to higher efficiency.
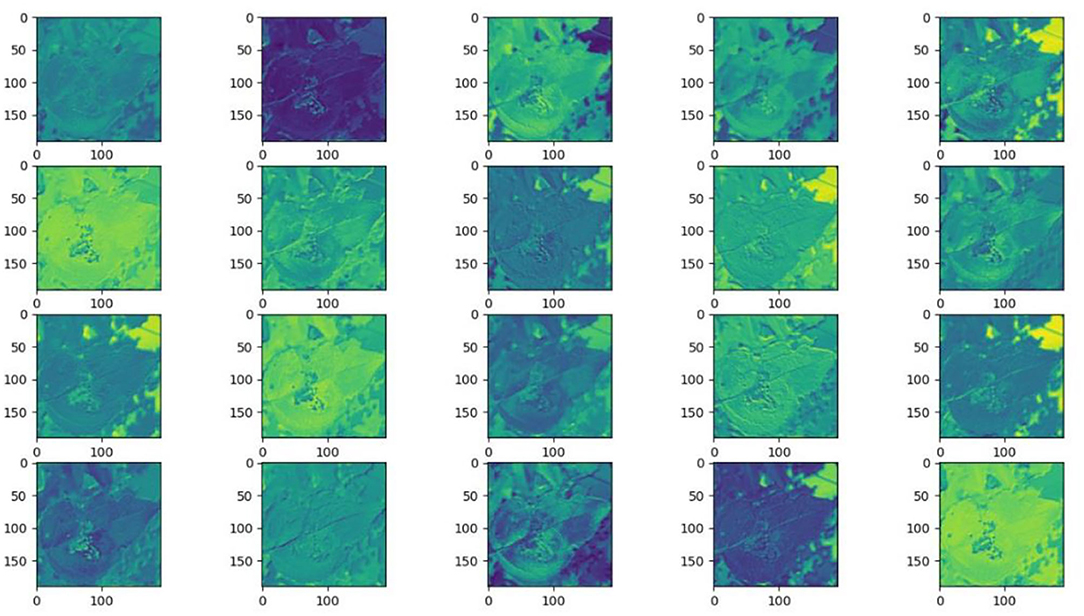
Figure 8 . Partial output feature maps of the first CA-Conv.
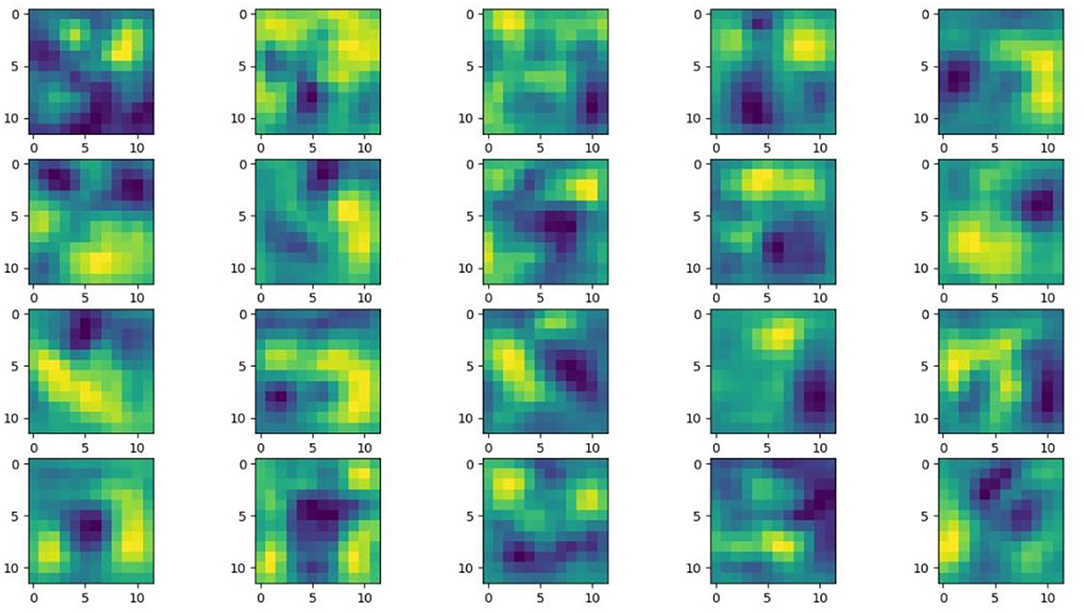
Figure 9 . Partial output feature maps of the last CA-Conv.
Class Activation Mapping (CAM) ( Selvaraju et al., 2020 ) helps to understand which feature components the model relies on to make decisions. Table 5 shows the original image of the class activation and the attention heatmaps of the commonly used models. The sample images of Glomerella leaf spot, black rot, apple litura moth, and rust are randomly selected for testing. Due to the introduction of the attention module CAB, CA-ENet has a stronger ability to focus on the lesion area. Compared with other models, CA-ENet has a good positioning effect and can accurately locate the interest area, whether it is a leaf lesion in a complex or a simple background. In contrast, ResNet-152, DenseNet-264, and ResNeXt-101 have deviations or even errors in their focus positions, which are what affect the robustness and accuracy of a model. The visual test results of the class activation heatmaps of the apple leaf diseases show that the model fully takes the characteristics of the disease spots into account and achieves superior recognition performance on apple leaf diseases.
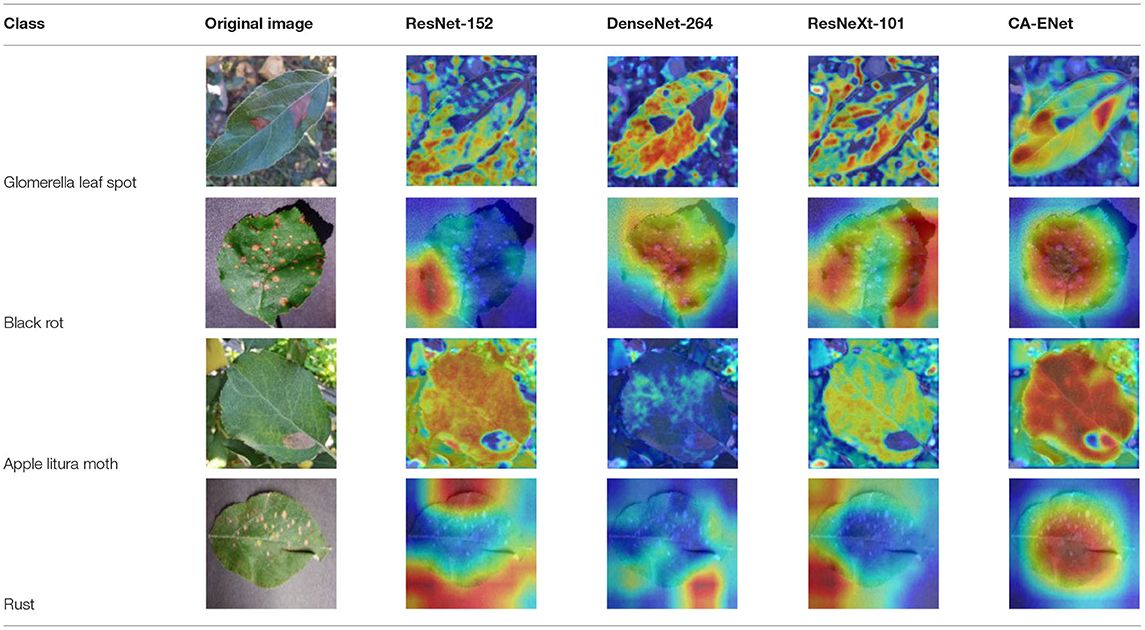
Table 5 . Comparison of attention heatmaps of different models.
Conclusion and Future Work
An improved attention-based deep CNN to identify common apple leaf diseases to support the efficient management of orchards is proposed in this study. Due to the complex environment of orchards, in order to be close to the real application scenarios, 5,170 apple leaf images were collected by multiple mobile devices and 3,000 disease images were obtained from a public dataset. Image augmentation techniques are used to generate the ALDID containing 81,700 diseased images. By embedding a CA block into a CA-Conv module, the integration of characteristic channel and location information was realized. A deep separable convolution is also used to reduce the number of parameters, and the h-swish activation function is used to speed up the model convergence. The proposed model is training with ALDID and testing with MRTD and conducts a large number of comparative experiments including various performance evaluation indicators and process visualizations. The experimental results show that the method proposed in this study achieves a recognition accuracy of 98.92%, which is better than that of other existing deep learning methods and achieves competitive performance on apple leaf disease identification tasks, which provides a reference for the application of deep learning methods in crop disease classification. The proposed model has the advantages of a simple structure, fast running speed, good generalization performance, and robustness, and has great potential application value. In the future, a ground mobile inspection platform equipped with cameras will be built to replace manual operations and to realize the rapid diagnosis and early warning of apple diseases.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
PW designed and performed the experiment, selected the algorithm, analyzed the data, trained the algorithms, and wrote the manuscript. PW, TN, YM, and ZZ collected data. BL monitored the data analysis. DH conceived the study and participated in its design. All authors contributed to the article and approved the submitted version.
This research is sponsored by the Key Research and Development Program of Shaanxi (2021NY-138 and 2019ZDLNY07-06-01), by CCF-Baidu Open Fund (NO. 2021PP15002000).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the reviewers for their valuable suggestions on this manuscript.
Aravind, K. R., Raja, P., Mukesh, K. V., Aniirudh, R., Ashiwin, R., and Szczepanski, C. (2018). “Disease classification in maize crop using bag of features and multiclass support vector machine,” in 2nd International Conference on Inventive Systems and Control (Coimbatore), 1191–1196. doi: 10.1109/ICISC.2018.8398993
CrossRef Full Text | Google Scholar
Bi, C., Wang, J., Duan, Y., Fu, B., Kang, J., and Shi, Y. (2020). MobileNet based apple leaf diseases identification. Mobile Netw. Appl . doi: 10.1007/s11036-020-01640-1
Chao, X., Sun, G., Zhao, H., Li, M., and He, D. (2020). Identification of apple tree leaf diseases based on deep learning models. Symmetry 12:1065. doi: 10.3390/sym12071065
Chollet, F. (2017). Xception: deep learning with depthwise separable convolutions. IEEE Conf. Comput. Vision Pattern Recogn . 1800–1807. doi: 10.1109/CVPR.2017.195
CrossRef Full Text
Dutot, M., Nelson, L., and Tyson, R. (2013). Predicting the spread of postharvest disease in stored fruit, with application to apples. Postharvest Biol. Technol. 85, 45–56. doi: 10.1016/j.postharvbio.2013.04.003
Guettari, N., Capelle-Laizé, A. S., and Carré, P. (2016). Blind image steganalysis based on evidential k-nearest neighbors. IEEE Int. Conf. Image Process . 2742–2746. doi: 10.1109/ICIP.2016.7532858
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep residual learning for image recognition. IEEE Conf. Comput. Vision Pattern Recogn . 770–778. doi: 10.1109/CVPR.2016.90
PubMed Abstract | CrossRef Full Text | Google Scholar
Hou, Q., Zhou, D., and Feng, J. (2021). Coordinate attention for efficient mobile network design. arXiv [Preprint]. arXiv:2103.02907v1.
Google Scholar
Howard, A., Sandler, M., Chen, B., Wang, W., Chen, L., Tan, M., et al. (2019). Searching for MobileNetV3. IEEE Int. Conf. Comput. Vision. 1314–1324. doi: 10.1109/ICCV.2019.00140
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., et al. (2017). Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv [Preprint]. arXiv:1704.04861.
Hu, J., Shen, L., Albanie, S., and Sun, G. (2018). Squeeze-and-excitation networks. IEEE Trans. Pattern. Anal . 7132–7141. doi: 10.1109/CVPR.2018.00745
Huang, G., Liu, Z., Maaten, L. V. D., and Weinberger, K. Q. (2017). Densely connected convolutional networks. IEEE Conf. Comput. Vision Pattern Recogn . 2261–2269. doi: 10.1109/CVPR.2017.243
Karthik, R., Hariharan, M., Anand, S., Mathikshara, P., Johnson, A., and Menaka, R. (2020). Attention embedded residual CNN for disease detection in tomato leaves. Appl. Soft Comput. 86:105933. doi: 10.1016/j.asoc.2019.105933
Kodovsky, J., Fridrich, J., and Holub, V. (2012). Ensemble classifiers for steganalysis of digital media. IEEE Trans. Inf. Foren Sec. 7, 432–444. doi: 10.1109/tifs.2011.2175919
Kour, V. P., and Arora, S. (2019). Particle swarm optimization based support vector machine (P-SVM) for the segmentation and classification of plants. IEEE Access 7, 29374–29385. doi: 10.1109/ACCESS.2019.2901900
Kulin, M., Kazaz, T., Moerman, I., and Poorter, E. D. (2018). End-to-end learning from spectrum data: a deep learning approach for wireless signal identification in spectrum monitoring applications. IEEE Access 6, 18484–18501. doi: 10.1109/ACCESS.2018.2818794
Liu, B., Ding, Z., Tian, L., He, D., Li, S., and Wang, H. (2020). Grape leaf disease identification using improved deep convolutional neural networks. Front. Plant Sci 11:1082. doi: 10.3389/fpls.2020.01082
Liu, B., Zhang, Y., He, D., and Li, Y. (2018). Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 10:11. doi: 10.3390/sym10010011
Ma, N., Zhang, X., Zheng, H. T., and Sun, J. (2018). Shufflenet v2: practical guidelines for efficient CNN architecture design. arXiv [Preprint]. arXiv:1807.11164v1.
Mi, Z., Zhang, X., Su, J., Han, D., and Su, B. (2020). Wheat stripe rust grading by deep learning with attention mechanism and images from mobile devices. Front. Plant Sci. 11:558126. doi: 10.3389/fpls.2020.558126
Mohammadpoor, M., Nooghabi, M. G., and Ahmedi, Z. (2020). An intelligent technique for grape fanleaf virus detection. Int. J. Interact. Multimedia Artif. Intell. 6, 62–67. doi: 10.9781/ijimai.2020.02.001
Prasad, S., Peddoju, S. K., and Ghosh, D. (2016). Multi-resolution mobile vision system for plant leaf disease diagnosis. Signal Image Video Process. 10, 379–388. doi: 10.1007/s11760-015-0751-y
Ramcharan, A., McCloskey, P., Baranowski, K., Mbilinyi, N., Mrisho, L., Ndalahwa, M., et al. (2019). A mobile-based deep learning model for cassava disease diagnosis. Front. Plant Sci. 10:272. doi: 10.3389/fpls.2019.00272
Ren, S., He, K., Girshick, R., and Sun, J. (2017). Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans. Pattern. Anal. 39, 1137–1149. doi: 10.1109/tpami.2016.2577031
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and Chen, L. (2018). MobileNetV2: inverted residuals and linear bottlenecks. IEEE Conf. Comput. Vision Pattern Recogn . 4510–4520. doi: 10.1109/CVPR.2018.00474
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., and Batra, D. (2020). “Grad-CAM: Visual Explanations From Deep Networks via Gradient-Based Localization,” in 2017 IEEE International Conference on Computer Vision (ICCV) , 618–626. doi: 10.1109/ICCV.2017.74
Sheikhan, M., Pezhmanpour, M., and Moin, M. S. (2012). Improved contourlet-based steganalysis using binary particle swarm optimization and radial basis neural networks. Neural Comput. Appl. 21, 1717–1728. doi: 10.1007/s00521-011-0729-9
Tan, M., and Quoc, V. L. (2019). EfficientNet: rethinking model scaling for convolutional neural networks. arXiv [Preprint]. arXiv:1905.11946.
Wang, X., Zhu, C., Fu, Z., Zhang, L., and Li, X. (2019). Research on cucumber powdery mildew recognition based on visual spectra. Spectrosc. Spectr. Anal. 39, 1864–1869. doi: 10.3964/j.issn.1000-0593 (2019)06-1864-06
Xie, S., Girshick, R., Dollár, P., Tu, Z., and He, K. (2017). Aggregated residual transformations for deep neural networks. IEEE Conf. Comput Vision Pattern. Recogn . 5987–5995. doi: 10.1109/CVPR.2017.634
Zhang, X., Zhou, X., Lin, M., and Sun, J. (2018a). ShuffleNet: an extremely efficient convolutional neural network for mobile devices. IEEE Conf. Comput. Vision Pattern. Recogn . 6848–6856. doi: 10.1109/CVPR.2018.00716
Zhang, Y., Gravina, R., Lu, H. M., Villari, M., and Fortino, G. (2018b). PEA: parallel electrocardiogram-based authentication for smart healthcare systems. J. Netw. Comput. Appl. 117, 10–16. doi: 10.1016/j.jnca.2018.05.007
Keywords: apple disease, CA-ENet, attention mechanism, CA block, diseases identification
Citation: Wang P, Niu T, Mao Y, Zhang Z, Liu B and He D (2021) Identification of Apple Leaf Diseases by Improved Deep Convolutional Neural Networks With an Attention Mechanism. Front. Plant Sci. 12:723294. doi: 10.3389/fpls.2021.723294
Received: 10 June 2021; Accepted: 23 August 2021; Published: 28 September 2021.
Reviewed by:
Copyright © 2021 Wang, Niu, Mao, Zhang, Liu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Liu, liubin0929@nwsuaf.edu.cn ; Dongjian He, hdj168@nwsuaf.edu.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Disease detection in apple leaves using deep convolutional neural network.

1. Introduction
2. related works.
- Previous models have limitations in utilising the advantage of Image Augmentation techniques properly. Our proposed model uses various Image Augmentation techniques such as Canny Edge Detection, Flipping, Blurring, etc., to enhance our dataset. These techniques can help in building a robust model.
- The performance of many of the previously proposed model were not adequate, especially under challenging cases, e.g., identifying leaves with multiple diseases. In our research, we have used an ensemble of pre-trained deep learning models. The advantage of this is that our proposed model combines the predictions of three models and can perform well under challenging situations.
- We have deployed the proposed model in the form of a web application that serves as an easy-to-use system for I users. The user has to upload the leaf’s image on our web application, and the result is obtained in a couple of seco.
3. Methodology
3.1. dataset collection, 3.1.1. healthy, 3.1.2. apple scab, 3.1.3. cedar apple rust, 3.1.4. multiple diseases, 3.2. image augmentation, 3.2.1. canny edge detection, 3.2.2. flipping, 3.2.3. convolution, 3.2.4. blurring, 3.3. dataset partition, 3.4. modeling, 3.4.1. multiclass classification, 3.4.2. transfer learning, 3.4.3. model structure, 3.4.4. densenet121, 3.4.5. efficientnet, 3.4.6. efficientnet noisy student, 3.5. ensembling, 4. experimental results and analyses, 4.1. experimental setup, 4.2. performance metrics, 4.3. performance benchmarking, 4.4. computational resources, 4.5. model deployment, 4.5.1. overview, 4.5.2. web application work flow.
- User visits our web application and uploads the image of apple leaf. All this takes place at the frontend.
- The image uploaded is then sent to the backend, where it is fed to the CNN model. At the backend, we have stored the weights of our proposed model in the form of an HDF5 file [ 57 ]. The model is loaded from this HDF5 file. Before feeding the image to the model, the image is first trimmed into the required shape of 512 × 512.
- The result returned by our model is a NumPy array of size (4,1), which includes the probability of the four classes. The class with the maximum probability is extracted.
- The results are shown to the user at the frontend, and the image is stored in our database to enhance our model.
4.5.3. Technologies Used
5. conclusions, author contributions, institutional review board statement, informed consent statement, data availability statement, acknowledgments, conflicts of interest.
- Apple. Wikipedia. 2021. Available online: https://en.wikipedia.org/wiki/Apple (accessed on 22 April 2021).
- Jordan, M.I.; Mitchell, T.M. Machine Learning: Trends, Perspectives, and Prospects. Science 2015 , 349 , 255–260. [ Google Scholar ] [ CrossRef ]
- Badage, A. Crop disease detection using machine learning: Indian agriculture. Int. Res. J. Eng. Technol. 2018 , 5 , 866–869. [ Google Scholar ]
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986 , PAMI-8 , 679–698. [ Google Scholar ] [ CrossRef ]
- Korkut, U.B.; Göktürk, Ö.B.; Yildiz, O. Detection of Plant Diseases by Machine Learning. In Proceedings of the 2018 26th Signal Processing and Communications Applications Conference (SIU), Izmir, Turkey, 2–5 May 2018; pp. 1–4. [ Google Scholar ]
- Noble, W.S. What Is a Support Vector Machine? Nat. Biotechnol. 2006 , 24 , 1565–1567. [ Google Scholar ] [ CrossRef ]
- Quinlan, J.R. Simplifying Decision Trees. Int. J. Man-Mach. Stud. 1987 , 27 , 221–234. [ Google Scholar ] [ CrossRef ] [ Green Version ]
- Huang, Y.; Li, L. Naive Bayes Classification Algorithm Based on Small Sample Set. In Proceedings of the 2011 IEEE International Conference on Cloud Computing and Intelligence Systems, Beijing, China, 15–17 September 2011; pp. 34–39. [ Google Scholar ]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015 , 521 , 436–444. [ Google Scholar ] [ CrossRef ]
- Kumar, E.P.; Sharma, E.P. Artificial neural networks-a study. Int. J. Emerg. Eng. Res. Technol. 2014 , 2 , 143–148. [ Google Scholar ]
- Pardede, H.F.; Suryawati, E.; Sustika, R.; Zilvan, V. Unsupervised Convolutional Autoencoder-Based Feature Learning for Automatic Detection of Plant Diseases. In Proceedings of the 2018 International Conference on Computer, Control, Informatics and Its Applications (IC3INA), Tangerang, Indonesia, 1–2 November 2018; pp. 158–162. [ Google Scholar ]
- Howard, A.G. Some Improvements on Deep Convolutional Neural Network Based Image Classification. arXiv 2013 , arXiv:1312.5402. [ Google Scholar ]
- Boulent, J.; Foucher, S.; Théau, J.; St-Charles, P.-L. Convolutional Neural Networks for the Automatic Identification of Plant Diseases. Front. Plant Sci. 2019 , 10 , 941. [ Google Scholar ] [ CrossRef ] [ Green Version ]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [ Google Scholar ]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 24 May 2019; pp. 6105–6114. [ Google Scholar ]
- Xie, Q.; Luong, M.-T.; Hovy, E.; Le, Q.V. Self-Training With Noisy Student Improves ImageNet Classification. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 10684–10695. [ Google Scholar ]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A Survey on Deep Transfer Learning. In Artificial Neural Networks and Machine Learning-ICANN 2018 Lecture Notes in Computer Science ; Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Springer: Cham, Germany, 2018; Volume 11141. [ Google Scholar ] [ CrossRef ] [ Green Version ]
- Steel, M.F.J. Model Averaging and Its Use in Economics. J. Econ. Lit. 2020 , 58 , 644–719. [ Google Scholar ] [ CrossRef ]
- Selvaraj, M.G.; Vergara, A.; Ruiz, H.; Safari, N.; Elayabalan, S.; Ocimati, W.; Blomme, G. AI-Powered Banana Diseases and Pest Detection. Plant Methods 2019 , 15 , 92. [ Google Scholar ] [ CrossRef ]
- Early Detection and Classification of Plant Diseases with Support Vector Machines Based on Hyperspectral Reflectance. Comput. Electron. Agric. 2010 , 74 , 91–99. [ CrossRef ]
- Sinha, A.; Shekhawat, R.S. Review of Image Processing Approaches for Detecting Plant Diseases. IET Image Process. 2019 , 14 , 1427–1439. [ Google Scholar ] [ CrossRef ]
- Ramesh, S.; Hebbar, R.; Niveditha, M.; Pooja, R.; Shashank, N.; Vinod, P.V. Plant Disease Detection Using Machine Learning. In Proceedings of the 2018 International Conference on Design Innovations for 3Cs Compute Communicate Control (ICDI3C), Bangalore, India, 25–28 April 2018; pp. 41–45. [ Google Scholar ]
- Yang, X.; Guo, T. Machine Learning in Plant Disease Research. Eur. J. Biomed. Res. 2017 , 3 , 6–9. [ Google Scholar ] [ CrossRef ] [ Green Version ]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016 , 7 . [ Google Scholar ] [ CrossRef ] [ PubMed ] [ Green Version ]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant Disease Detection and Classification by Deep Learning. Plants 2019 , 8 , 468. [ Google Scholar ] [ CrossRef ] [ Green Version ]
- Goncharov, P.; Ososkov, G.; Nechaevskiy, A.; Uzhinskiy, A.; Nestsiarenia, I. Disease Detection on the Plant Leaves by Deep Learning. In Proceedings of the International Conference on Neuroinformatics, Moscow, Russia, 8–12 October 2018; Kryzhanovsky, B., Dunin-Barkowski, W., Redko, V., Tiumentsev, Y., Eds.; Advances in Neural Computation, Machine Learning, and Cognitive Research II. Springer International Publishing: Cham, Germany, 2019; pp. 151–159. [ Google Scholar ]
- Sun, J.; Yang, Y.; He, X.; Wu, X. Northern Maize Leaf Blight Detection Under Complex Field Environment Based on Deep Learning. IEEE Access 2020 , 8 , 33679–33688. [ Google Scholar ] [ CrossRef ]
- Vine Disease Detection in UAV Multispectral Images Using Optimized Image Registration and Deep Learning Segmentation Approach. Comput. Electron. Agric. 2020 , 174 , 105446. [ CrossRef ]
- Identification of Rice Diseases Using Deep Convolutional Neural Networks. Neurocomputing 2017 , 267 , 378–384. [ CrossRef ]
- Amara, J.; Bouaziz, B.; Algergawy, A. A Deep Learning-Based Approach for Banana Leaf Diseases Classification. In Datenbanksysteme für Business, Technologie und Web (BTW 2017)-Workshopband ; Mitschang, B., Nicklas, D., Leymann, F., Schöning, H., Herschel, M., Teubner, J., Härder, T., Kopp, O., Wieland, M., Eds.; Gesellschaft für Informatik e.V.: Bonn, Germany, 2017; pp. 79–88. [ Google Scholar ]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998 , 86 , 2278–2324. [ Google Scholar ] [ CrossRef ] [ Green Version ]
- Factors Influencing the Use of Deep Learning for Plant Disease Recognition. Biosyst. Eng. 2018 , 172 , 84–91. [ CrossRef ]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 2018 , 10 , 11. [ Google Scholar ] [ CrossRef ] [ Green Version ]
- Chuanlei, Z.; Shanwen, Z.; Jucheng, Y.; Yancui, S.; Jia, C. Apple Leaf Disease Identification Using Genetic Algorithm and Correlation Based Feature Selection Method. Int. J. Agric. Biol. Eng. 2017 , 10 , 74–83. [ Google Scholar ] [ CrossRef ]
- Dai, B.; Qiu, T.; Ye, K. Foliar Disease Classification. Available online: http://noiselab.ucsd.edu/ECE228/projects/Report/15Report.pdf (accessed on 20 April 2021).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [ Google Scholar ]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper With Convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [ Google Scholar ]
- Sardoğan, M.; Özen, Y.; Tuncer, A. Detection of Apple Leaf Diseases Using Faster R-CNN. Düzce Üniversitesi Bilim Teknol. Derg. 2020 , 8 , 1110–1117. [ Google Scholar ] [ CrossRef ]
- Jiang, P.; Chen, Y.; Liu, B.; He, D.; Liang, C. Real-Time Detection of Apple Leaf Diseases Using Deep Learning Approach Based on Improved Convolutional Neural Networks. IEEE Access 2019 , 7 , 59069–59080. [ Google Scholar ] [ CrossRef ]
- Jeong, J.; Park, H.; Kwak, N. Enhancement of SSD by Concatenating Feature Maps for Object Detection. arXiv 2017 , arXiv:1705.09587. [ Google Scholar ]
- Plant Pathology 2020-FGVC7. Available online: https://kaggle.com/c/plant-pathology-2020-fgvc7/data/ (accessed on 20 April 2021).
- Perez, L.; Wang, J. The Effectiveness of Data Augmentation in Image Classification Using Deep Learning. arXiv 2017 , arXiv:1712.04621. [ Google Scholar ]
- Glossary-Convolution. Available online: https://homepages.inf.ed.ac.uk/rbf/HIPR2/convolve.htm (accessed on 18 April 2021).
- OpenCV: Smoothing Images. Available online: https://docs.opencv.org/master/d4/d13/tutorial_py_filtering.html (accessed on 18 April 2021).
- Lin, M.; Chen, Q.; Yan, S. Network In Network. arXiv 2014 , arXiv:1312.4400. [ Google Scholar ]
- Nwankpa, C.; Ijomah, W.; Gachagan, A.; Marshall, S. Activation Functions: Comparison of Trends in Practice and Research for Deep Learning. arXiv 2018 , arXiv:1811.03378. [ Google Scholar ]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017 , arXiv:1412.6980. [ Google Scholar ]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [ Google Scholar ]
- Nain, A. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. Available online: https://medium.com/@nainaakash012/efficientnet-rethinking-model-scaling-for-convolutional-neural-networks-92941c5bfb95 (accessed on 25 May 2021).
- Ju, C.; Bibaut, A.; van der Laan, M. The Relative Performance of Ensemble Methods with Deep Convolutional Neural Networks for Image Classification. J. Appl. Stat. 2018 , 45 , 2800–2818. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Kaggle: Your Machine Learning and Data Science Community. Available online: https://www.kaggle.com/ (accessed on 20 April 2021).
- Tensor Processing Units (TPUs) Documentation. Available online: https://www.kaggle.com/docs/tpu (accessed on 20 April 2021).
- Tf.Data.Dataset | TensorFlow Core v2.4.1. Available online: https://www.tensorflow.org/api_docs/python/tf/data/Dataset (accessed on 20 April 2021).
- Seaborn: Statistical Data Visualization—Seaborn 0.11.1 Documentation. Available online: https://seaborn.pydata.org/#:~:text=Seaborn%20is%20a%20Python%20data,attractive%20and%20informative%20statistical%20graphics (accessed on 20 April 2021).
- Mishra, A. Metrics to Evaluate Your Machine Learning Algorithm. Available online: https://towardsdatascience.com/metrics-to-evaluate-your-machine-learning-algorithm-f10ba6e38234 (accessed on 18 April 2021).
- Available online: https://github.com/prakhar070/apple-disease-detection (accessed on 20 April 2021).
- The HDF5® Library & File Format. The HDF Group. Available online: https://www.hdfgroup.org/solutions/hdf5/ (accessed on 22 April 2021).
| Split | DenseNet121 | EfficientNetB7 | NoisyStudent | Our Model |
|---|---|---|---|---|
| 0.10 | 0.9398 | 0.9416 | 0.9095 | 0.9344 |
| 0.15 | 0.9526 | 0.9562 | 0.9124 | 0.9625 |
| 0.20 | 0.9511 | 0.9506 | 0.9233 | 0.9499 |
| 0.25 | 0.9414 | 0.9471 | 0.8991 | 0.9211 |
| 0.30 | 0.9232 | 0.9376 | 0.9091 | 0.9301 |
| Split | DenseNet121 | EfficientNetB7 | NoisyStudent | Our Model |
|---|---|---|---|---|
| 0.10 | 0.8432 | 0.8637 | 0.8344 | 0.8555 |
| 0.15 | 0.8637 | 0.8901 | 0.8479 | 0.9091 |
| 0.20 | 0.9496 | 0.8871 | 0.8544 | 0.8963 |
| 0.25 | 0.8598 | 0.8388 | 0.8282 | 0.8446 |
| 0.30 | 0.8876 | 0.8876 | 0.8298 | 0.8991 |
| Split | DenseNet121 | EfficientNetB7 | NoisyStudent | Our Model |
|---|---|---|---|---|
| 0.10 | 0.8442 | 0.8637 | 0.8344 | 0.8518 |
| 0.15 | 0.8637 | 0.8961 | 0.8429 | 0.8977 |
| 0.20 | 0.9496 | 0.8870 | 0.8542 | 0.8951 |
| 0.25 | 0.8221 | 0.8388 | 0.8238 | 0.8331 |
| 0.30 | 0.8806 | 0.8806 | 0.8301 | 0.8881 |
| Split | DenseNet121 | EfficientNetB7 | NoisyStudent | Our Model |
|---|---|---|---|---|
| 0.10 | 0.8442 | 0.8637 | 0.8329 | 0.8497 |
| 0.15 | 0.8637 | 0.8922 | 0.8453 | 0.9098 |
| 0.20 | 0.9496 | 0.8871 | 0.8576 | 0.8732 |
| 0.25 | 0.8560 | 0.8382 | 0.8249 | 0.8490 |
| 0.30 | 0.8839 | 0.8839 | 0.8290 | 0.8987 |
| Model | Accuracy |
|---|---|
| DenseNet | 0.9260 |
| GoogleNet | 0.9530 |
| EfficientNetB7 | 0.9562 |
| ResNet20 | 0.9370 |
| VggNet-16 | 0.9400 |
| Our Model | 0.9625 |
| MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
Share and Cite
Bansal, P.; Kumar, R.; Kumar, S. Disease Detection in Apple Leaves Using Deep Convolutional Neural Network. Agriculture 2021 , 11 , 617. https://doi.org/10.3390/agriculture11070617
Bansal P, Kumar R, Kumar S. Disease Detection in Apple Leaves Using Deep Convolutional Neural Network. Agriculture . 2021; 11(7):617. https://doi.org/10.3390/agriculture11070617
Bansal, Prakhar, Rahul Kumar, and Somesh Kumar. 2021. "Disease Detection in Apple Leaves Using Deep Convolutional Neural Network" Agriculture 11, no. 7: 617. https://doi.org/10.3390/agriculture11070617
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals

- Advanced Search
Research on deep learning in apple leaf disease recognition
College of Information and Electrical Engineering, China Agricultural University, Beijing 100083, China
- 13 citation
New Citation Alert added!
This alert has been successfully added and will be sent to:
You will be notified whenever a record that you have chosen has been cited.
To manage your alert preferences, click on the button below.
New Citation Alert!
Please log in to your account
- Publisher Site
Computers and Electronics in Agriculture

• | Use DenseNet-121 as the backbone network. | ||||
• | Three methods were proposed to identify apple leaf diseases. | ||||
• | The recognition accuracy is improved compared with the traditional method. | ||||
The main reason affecting apple production is the occurrence of apple leaf diseases, which causes huge economic losses every year. Therefore, it is of great significance to study the identification of apple leaf diseases. Based on DenseNet-121 deep convolution network, three methods of regression, multi-label classification and focus loss function were proposed to identify apple leaf diseases. In this paper, the apple leaf image data set, including 2462 images of six apple leaf diseases, were used for data modeling and method evaluation. The proposed methods achieved 93.51%, 93.31% and 93.71% accuracy on the test set respectively, which were better than the traditional multi-classification method based on cross-entropy loss function with an accuracy of 92.29%.
Index Terms
Applied computing
Life and medical sciences
Computing methodologies
Artificial intelligence
Computer vision
Machine learning
Learning paradigms
Supervised learning
Machine learning approaches
Neural networks
Recommendations
Mobilenet based apple leaf diseases identification.
Alternaria leaf blotch, and rust are two common types of apple leaf diseases that severely affect apple yield. A timely and effective detection of apple leaf diseases is crucial for ensuring the healthy development of the apple industry. In ...
Leaf disease detection using machine learning and deep learning: Review and challenges
Identification of leaf disorder plays an important role in the economic prosperity of any country. Many parts of a plant can be infected by a virus, fungal, bacteria, and other infectious organisms but here we mainly considered the ...
- A study on leaf disease (LD) detection is conducted using Machine Learning and Deep Learning from 2010 to 2022.
Leaf Disease Classification of Various Crops Using Deep Learning Based DBESeriesNet Model
Chilli is one of the world’s most extensively used spices. Numerous diseases affect chilli leaves as a result of climatic and environmental changes, which limit crop output. Chilli leaf disease drastically reduces the quality and yield of the ...
Login options
Check if you have access through your login credentials or your institution to get full access on this article.
Full Access
- Information
- Contributors
Published in
Elsevier B.V.
In-Cooperation
Elsevier Science Publishers B. V.
Netherlands
Publication History
- Published: 1 January 2020
Author Tags
- Deep learning
- Apple leaves
- Disease recognition
- Multi-label classification
- Focus loss function
- Cross entropy loss function
- research-article
Funding Sources
Other metrics.
- Bibliometrics
- Citations 13
Article Metrics
- 13 Total Citations View Citations
- 0 Total Downloads
- Downloads (Last 12 months) 0
- Downloads (Last 6 weeks) 0
Digital Edition
View this article in digital edition.
Share this Publication link
https://dl.acm.org/doi/10.1016/j.compag.2019.105146
Share on Social Media
- 0 References
Export Citations
- Please download or close your previous search result export first before starting a new bulk export. Preview is not available. By clicking download, a status dialog will open to start the export process. The process may take a few minutes but once it finishes a file will be downloadable from your browser. You may continue to browse the DL while the export process is in progress. Download
- Download citation
- Copy citation
We are preparing your search results for download ...
We will inform you here when the file is ready.
Your file of search results citations is now ready.
Your search export query has expired. Please try again.
Click through the PLOS taxonomy to find articles in your field.
For more information about PLOS Subject Areas, click here .
Loading metrics
Open Access
Peer-reviewed
Research Article
A method of detecting apple leaf diseases based on improved convolutional neural network
Roles Writing – original draft
Affiliation Center for Innovation Management Research of Xinjiang, School of Economics and Management, Xinjiang University, Urumqi, Xinjiang, China
Roles Supervision, Writing – review & editing
* E-mail: [email protected]
- Jie Di,

- Published: February 1, 2022
- https://doi.org/10.1371/journal.pone.0262629
- Reader Comments
Apple tree diseases have perplexed orchard farmers for several years. At present, numerous studies have investigated deep learning for fruit and vegetable crop disease detection. Because of the complexity and variety of apple leaf veins and the difficulty in judging similar diseases, a new target detection model of apple leaf diseases DF-Tiny-YOLO, based on deep learning, is proposed to realize faster and more effective automatic detection of apple leaf diseases. Four common apple leaf diseases, including 1,404 images, were selected for data modeling and method evaluation, and made three main improvements. Feature reuse was combined with the DenseNet densely connected network and further realized to reduce the disappearance of the deep gradient, thus strengthening feature propagation and improving detection accuracy. We introduced Resize and Re-organization (Reorg) and conducted convolution kernel compression to reduce the calculation parameters of the model, improve the operating detection speed, and allow feature stacking to achieve feature fusion. The network terminal uses convolution kernels of 1 × 1, 1 × 1, and 3 × 3, in turn, to realize the dimensionality reduction of features and increase network depth without increasing computational complexity, thus further improving the detection accuracy. The results showed that the mean average precision (mAP) and average intersection over union (IoU) of the DF-Tiny-YOLO model were 99.99% and 90.88%, respectively, and the detection speed reached 280 FPS. Compared with the Tiny-YOLO and YOLOv2 network models, the new method proposed in this paper significantly improves the detection performance. It can also detect apple leaf diseases quickly and effectively.
Citation: Di J, Li Q (2022) A method of detecting apple leaf diseases based on improved convolutional neural network. PLoS ONE 17(2): e0262629. https://doi.org/10.1371/journal.pone.0262629
Editor: Nguyen Quoc Khanh Le, Taipei Medical University, TAIWAN
Received: July 15, 2021; Accepted: December 31, 2021; Published: February 1, 2022
Copyright: © 2022 Di, Li. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: All data files are available from the PlantVillage database ( https://plantvillage.org/ ). These are third party data (i.e., data not owned or collected by the author(s)) and we confirm that we did not have any special access privileges that others would not have.
Funding: National Natural Science Foundation of China (82060265)” and “the Department of Human Resources and Social Security of Xinjiang Uygur Autonomous Region (042419001).
Competing interests: The authors have declared that no competing interests exist.
1. Introduction
With over 2,000 years of history in China, areas of apple implantation have expanded annually, and 65% of all apples in the world are now produced in China. The traditional apple industry has been modernized with the support of national economic policies, and China has gradually developed into a major power source in the apple industry. Nonetheless, behind the rapid development of the apple implantation industry, disease prevention and control have been important problems that have long perplexed orchard farmers [ 1 ], who identify apple diseases by referring to experience, books, the Internet, and consulting professional and technical personnel [ 2 ]. However, relying solely on these sources is not conducive to timely and effective identification of diseases, and might even cause other problems associated with subjective judgment. Effective automated detection of apple diseases during production not only promptly monitors the health status of apples but also helps orchard farmers to correctly judge apple diseases. They can then implement timely prevention and control to avoid large-scale diseases, which is crucial for promoting the healthy growth of apples and increasing the economic benefits of orchards [ 3 , 4 ].
Hundreds of apple diseases disseminate in fruits, leaves, branches, roots, and other areas, but often initially appear in leaves, which are easily observed, collected, and managed. Therefore, they are an important reference for disease identification and effective automated detection of diseases is essential. However, judging differences among diseases is difficult due to the complexity of blade veins [ 5 ], resulting in unsatisfactory outcomes of experimental detection methods [ 6 ].
Nowadays, based on traditional algorithms, much methods has been made in the detection of apple leaf diseases, artificial neural network algorithms, genetic algorithms [ 7 ], support vector machines (SVMs), and a clustering algorithm [ 8 ] have applied feature extraction to apple leaves, and Li and He [ 9 ] established a BP network model with intelligent recognition that can diagnose apple leaf diseases using low-resolution images. Rong-Ming, Wei [ 10 ] proposed a method for extracting apple leaf disease spots based on hyperspectral images. Omrani, Khoshnevisan [ 11 ] proposed a calculation based on an artificial neural network (ANN) and SVMs to detect three apple leaf diseases. Shuaibu, Lee [ 12 ] applied hyperspectral images to detect marssonina blotch in apples at different stages and used orthogonal subspace projection for feature selection and redundancy reduction which generated better results. Zhang, Zhang [ 7 ] used genetic algorithm and feature selection method based on correlation to identify three kinds of apple leaf diseases, with an average recognition rate of 90%. Although traditional methods can also extract a large number of classification features, to select a few optimal features using general feature selection methods is difficult, so the recognition rate of these methods is low and generalization ability is weak.
Since the emergence of target detection technology during the 1960s, many domestic and overseas investigators have focused on the development of new technologies. After Krizhevsky, Sutskever [ 13 ] proposed the AlexNet structure at the ImageNet Large-scale Visual Recognition Challenge (ILSVRC) in 2012, target detection based on deep learning has gradually become mainstream [ 14 ]. Instead of using manual feature extraction, deep learning adopts hierarchical feature extraction [ 15 ], which is more similar to the human visual mechanism and has more accurate detection [ 16 ]. Many investigations into target detection based on deep learning have emerged [ 17 ], and applications to fruit and vegetable crop disease detection have been continuously proposed. Based on the AlexNet model, Brahimi, Boukhalfa [ 18 ] classified nine tomato diseases using more than 10,000 tomato disease images with a simple background in the PlantVillage database, further enhancing the model recognition rate. Priyadharshini, Arivazhagan [ 19 ] proposed a deep CNN-based architecture (modified LeNet) for four maize leaf disease classifications, and the trained model achieved 97.89% accuracy. In order to effectively examine the minor disease spots on diseased grape leaves, Zhu, Cheng [ 20 ] proposed a detection algorithm based on convolutional neural network and super-resolution image enhancement, the test effect is greatly improved. Zhang, Zhang [ 21 ] improved the model structure of the LeNet convolutional neural network to solve the problems of leaf shape, illumination, and image background that easily affect detection using traditional recognition methods; their model recognized six cucumber diseases, with an average recognition ratio of 90.32%. Xue, Huang [ 22 ], overcame the difficulty of detecting mango overlap or occlusion and proposed the improved Tiny-YOLO mango detection network combining dense connections, with an accuracy rate of 97.02%. Xiao, Li-ren [ 23 ] proposed the feature pyramid networks-single-shot multibox detector (FPN-SSD) model to detect top soybean leaflets on collected images, and automatically detect soybean leaves and classify leaf morphology. The wide application of deep learning to the detection of fruit and vegetable diseases has furthered automatic disease detection by leaps and bounds [ 24 , 25 ].
Convolutional neural network learning methods are now being applied to classify and identify apple leaf diseases, such as the SDD model, and the R-CNN and YOLO series of algorithms. Many improved schemes have been proposed based on existing algorithms. Jiang, Chen [ 26 ] proposed the INAR-SSD (SSD with an Inception module and Rainbow concatenation) model to detect five common apple leaf diseases, with high accuracy (78.80% mAP) in real time. The INAR-SSD has provided feasible real-time detection of apple leaf diseases. Liu, Zhang [ 27 ] created a new structure based on the deep convolutional neural network AlexNet to detect apple leaf diseases, thus further improving the effective identification of apple leaf diseases. Based on the DenseNet-121 deep convolutional network, Zhong and Zhao [ 28 ] proposed regression, multi-label classification, and focus loss function, to identify apple leaf diseases, with a recognition accuracy rate > 93%, which was better than the traditional multi-classification method based on the cross-entropy loss function. Jan and Ahmad [ 29 ] developed an apple pest and disease diagnostic system to predict apple scabs and leaf spots. Entropy, energy, inverse difference moment (IDM), mean, standard deviation (SD), and perimeter, etc., were extracted from apple leaf images and a multi-layer perceptron (MLP) pattern classifier and 11 apple leaf image features to train the model, which had 99.1% diagnostic accuracy. Yu, Son [ 30 ] proposed a new method based on a region-of-interest-aware deep convolutional neural network (ROI-aware DCNN) to render deep features more discriminative and increase classification performance for apple leaf disease identification, with an average accuracy of 84.3%. Sun, Xu [ 31 ] proposed a lightweight CNN model that can be deployed on mobile devices to detect apple leaf diseases in real time to test five common apple leaf diseases, with a mAP value of 83.12%. Di and Qu [ 32 ] applied the Tiny-YOLO model to apple leaf diseases, and the results showed that the model was effective.
There are two main types of target detection in the deep learning era [ 33 ]. The first is based on region formation. Representative models are R-CNN [ 34 ], Fast R-CNN [ 35 ], and Faster R-CNN [ 36 ]. One method forms candidate boxes that might contain target areas in images, extracts features from each box, trains classifiers, and achieves the target detection of specific objects in two steps. The other method is based on regression. Typical representative models are SDD [ 37 ], YOLO [ 38 ], and YOLOv2 [ 39 ]. These methods address detection as regression problems; they directly predict the coordinates of the boxes, the confidence level of the objects contained, and the probability of the objects appearing, then realizes the target location and identifies the category of specific objects in one step. The detection accuracy of the region formation-based method is superior. However, a target detection method based on regression is more advantageous in terms of detection speed and is popular in terms of real-time detection and intelligent applications. Yet, most recent research efforts focusing on detecting apple leaf diseases are not enough, and considerable room exists for improving detection accuracy and effectiveness. Therefore, the present study investigated how to optimize methods of apple leaf disease detection. Practical problems such as complexity and variations in the veins of apple leaves and difficulties with disease identification can be overcome by the rapid and effective automated detection of apple leaf diseases. Therefore, we created the DF-Tiny-YOLO model to, an detect apple leaf diseases based on regression with the concepts of DenseNet [ 40 ] and F-YOLO [ 41 ]. We then optimized the model considering rapid, accurate detection.
2. Algorithm principle and DF-Tiny-YOLO network
2.1 algorithm principles, 2.1.1 yolo network model..
On this basis, YOLO selects the appropriate candidate region to predict while detecting the input picture to obtain the optimal position and classification of objects in the candidate region. The YOLO network model comprises 24 convolutions and two full-link layers. Redmon, Divvala [ 38 ] applied the YOLO algorithm to various datasets to verify its accuracy. The average YOLO target detection accuracy is about 60%, with a recognition rate up to 45 frames/s.
2.1.2 YOLOv2 network.
Although YOLO has been greatly improved in terms of detection speed, its detection accuracy is far from optimal. Object positioning is insufficiently accurate, and its recall rate is relatively low. Thus, YOLOv2 has been improved based on the advantages of the original YOLO, which proposes the new network structure, Darenet-19, which includes 19 convolution and five maximum pooling layers and adopts a series of optimization strategies. In terms of candidate box selection, the K-means clustering method is adopted in YOLOv2 to determine the size, proportion, and number of candidate boxes, and obtain reasonable candidate box selection parameters through balanced clustering results and computational complexity. A pass-through layer has been added to the network structure to connect shallow and deep features, which has adapted YOLOv2 to multi-scale features and improved the detection accuracy of small targets. Multi-scale training was adopted, and a group of new pictures was randomly selected for scale input every 10 rounds of training to enhance detection performance. Moreover, many aspects have been improved. The final test results of the VOC 2007 dataset showed that when the size of the input picture was 228 × 228, the frame rate reached 67 FPS and mAP reached 76.8%. When the input picture resolution was 554 × 554, the mAP of YOLOv2 on VOC 2007 reached 78.6% and the frame rate reached 40 FPS, which satisfied real-time requirements.
2.1.3 Tiny-YOLO network.
Tiny-YOLO is a miniature “accelerated version” network based on the Darknet-19 structure ( Fig 1 ). This reduces the complexity of the model, and the number of training parameters greatly improves the training speed up to 244 frames/s, which is 1.5-fold the detection speed of YOLOv2. However, noise increased during Tiny-YOLO training due to reduced model depth. Despite progress in speed, target positioning accordingly became inaccurate, resulting in a low average accuracy of 23.7%, compared with YOLOv2, which can achieve 78.6% accuracy at 40 frames/s. The detection accuracy of Tiny-YOLO can still be improved.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
Tiny-YOLO structure and the network parameter shown in here.
https://doi.org/10.1371/journal.pone.0262629.g001

2.1.4 Dense connective network.
Huang, Liu [ 40 ] connected all layers in the network in pairs so that each layer in the network can receive the characteristics of all layers in front of it as input. This maximized information flow among all layers in the network. This network structure is called DenseNet because of its numerous dense connections ( Fig 2 ).
https://doi.org/10.1371/journal.pone.0262629.g002
DenseNet has the following important characteristics. It alleviates the problem of gradient dissipation during training to some extent. Each layer receives gradient signals from all following layers during reverse transmission, and the gradient near the input layer will not become smaller as the network depth increases. As numerous features are reused, many features can be generated using fewer convolution kernels, so the size of the final model is relatively small. The main purpose of DenseNet was to establish a connection relationship between different layers. In the traditional convolution structure, layer k has k connections, whereas in the DenseNet structure, layer k has k ( k − 1)/2 connections, that is, each layer in the model is connected with the following layer to enhance feature extraction. Fig 3 shows the network structure of DenseNet. Overall, the structure consists mainly of dense blocks and transition layers. The internal feature diagrams of the dense blocks are of the same size, and transition block nodes comprise “BN-Conv-Pool”.
https://doi.org/10.1371/journal.pone.0262629.g003
DenseNet encourages feature reuse, which means that the features of each picture can be used, and this reduces disappearance of feature gradient, strengthens the feature propagation of the network model, and effectively inhibits the overfitting problem.
2.1.5 F-YOLO network.
Mehta and Ozturk [ 41 ] proposed F-YOLO based on Tiny-YOLO. Generally, a deeper and wider network structure, results in a better detection effect, but the corresponding calculation of parameters will also increase, resulting in slow network training. Considering this problem, Mehta et al. used the resize in the YOLOv2 algorithm to compress the convolution layers of the 6th, 8th, and 10th layers by a 1 × 1 convolution kernel, superimpose them on the first 13 × 13 × 1024 convolution layer, and connect them to form a 13 × 13 × 1792 feature diagram. Thereafter, many 1 × 1 convolution layers were added to increase the network depth without increasing the computational complexity, thus producing a “narrow and deep” network. Compared with the original Tiny-YOLO network, the modified network improved the accuracy by 5 mAP and the speed by 20%. On the Pascal dataset, the training parameters of the final model were reduced > 10-fold, and the training speed exceeded 200 FPS compared with the VGG-based target detection model.
2.2 DF-Tiny-YOLO network model
The following improvements to the original Tiny-YOLO network structure were made based on existing knowledge to realize the rapid identification and accurate location of apple leaf diseases.
Huang, Liu [ 40 ] proposed the concept of a dense connection network. They replaced the 13 × 13 × 512 convolution layer in the Tiny-YOLO structure with a dense module; the 13 × 13 × 128 convolution kernel was used for convolution to output 128 feature diagrams, so that the spliced output of the input and first layers is 13 × 13 × (512 + 128), the output set of the second layer is 13 × 13 × (640 + 128), and so on. The spliced output of the last layer is 13 × 13 × 1024, thus reducing gradient disappearance, and further realizing feature reuse, strengthening feature propagation, and improving detection accuracy.
Resize and reorg were adopted to compress large feature diagrams, reduce network computation, accelerate network training, and improve the speed of detection. Large and small feature diagrams were stacked on the last main convolution layer to achieve feature fusion.
Using 1 × 1, 1 × 1, and 3 × 3 convolution kernels at the network terminals will contribute to reducing the dimension of features and increasing the network depth without increasing computational complexity, thus further improving the detection accuracy.
A DF-Tiny-YOLO network model was constructed with the structure shown in Fig 3 based on these improvements, and the network parameter also shown in here.
3. Analysis of experimental process and results
3.1 experimental process, 3.1.1 experimental data..
We randomly selected four clear common apple leaf pictures of the same size in the PlantVillage database as the experimental dataset. The quality of this dataset was ensured by manual screening to avoid the problems of singleness, repeatability, and errors. The data included 300 images of leaves with apple scab, 300 with apple black rot, 275 with apple cedar rust, and 529 healthy leaves, total 1,404 images. Fig 4 describes the basic situation of four types of leaves. The images were randomly divided into training and test sets at a ratio of 80%:20%, the division of data set was shown in Table 1 .
The figure describes the basic situation of four types of leaves.
https://doi.org/10.1371/journal.pone.0262629.g004
https://doi.org/10.1371/journal.pone.0262629.t001
Many common diseases affect apple leaves. Table 2 summarizes the main characteristics of diseased and healthy leaves with reference to relevant information.
https://doi.org/10.1371/journal.pone.0262629.t002
After uniformly labeling the four categories of apple leaf picture data, the leaf samples were manually labeled individually according to the following criteria: label box frames apple leaves in the image regardless of the stem, and the upper, lower, left, and right sides of the label box are close to the outer boundary of the leaves, to minimize errors emerging during sample labeling.
3.1.2 Environment and tools.
Table 3 shows the server platform configuration and tools used herein.
https://doi.org/10.1371/journal.pone.0262629.t003
3.1.3 Model training.
Referring to the training parameters of the original Tiny-YOLO model, the main initial training parameters of the improved DF-Tiny-YOLO model set in this study are shown in the Table 4 . Where the learning rate parameter values will be adjusted appropriately during the model training period based on the average loss values.
https://doi.org/10.1371/journal.pone.0262629.t004
Fig 5 shows the experimental process for the target detection of apple leaf diseases. The apple leaf images are first screened, then the entire leaf data set is divided into a training set and a test set, and finally an experimental file containing the corresponding information is generated by annotating the samples. Using the apple leaf training data, the detection training is performed on the original YOLOv2 model, the Tiny-YOLO model, and the modified DF-Tiny-YOLO model, respectively. The DF-Tiny-YOLO network model was generated by improving the Tiny-YOLO network through combining DenseNet with the F-YOLO concept. The model was trained and tested using the training and test datasets, respectively, since the measurement error decreases in many iterations, the network performance is optimized with the correction of parameters. The training ends when the average loss value of the network model decreases to a certain level and the value does not decrease in subsequent iterations, to generate the DF-Tiny-YOLO apple leaf disease detection model. The experimental findings of DF-Tiny-YOLO, YOLOv2, and the original Tiny-YOLO were compared.
The figure shows the experimental process for the target detection of apple leaf diseases.
https://doi.org/10.1371/journal.pone.0262629.g005
3.1.4 Experimental evaluation indexes.
The mean average precision (mAP), intersection over union (IoU), recall, and frame per second (FPS) were adopted as the evaluation indexes of the model to detect apple leaf diseases. The results were assessed as true positive (TP), true negative (TN), false positive (FP), and false negative (FN).
Frame Rate per Second (FPS) is a common indicator of speed, namely, the number of pictures that can be processed per second.
3.2 Experimental results and analysis
To prevent the prediction model from overfitting, we determine whether the training model is optimal by observing the average loss value. We end the training when the average loss value decreases to a certain level and the value does not decrease again in the subsequent iterations. The training is stopped when the average loss of the network model decreases to the level of 0.0048. Fig 6 shows variations in the average losses of DF-Tiny-YOLO and Tiny-YOLO during training. Generally, the model rapidly fits at the start of the iteration, and the loss also rapidly decreases. With the continuous increase in the number of iterations, the loss gradually tended to stabilize during forward and backward oscillation. The loss values of DF-Tiny-YOLO and Tiny-YOLO in the first iteration were 23.79% and 28.46%, respectively. As the number of iterations increased, the loss values decreased faster in DF-Tiny-YOLO than Tiny-YOLO. The model fitting was faster, the oscillation amplitude is more moderate, and the final stationary value was relatively smaller. These results show that the proposed DF-Tiny-YOLO model has better training effects.
The figure shows variations in the average losses of DF-Tiny-YOLO and Tiny-YOLO during training.
https://doi.org/10.1371/journal.pone.0262629.g006
Table 5 compares the experimental results of DF-Tiny-YOLO, YOLOv2, and Tiny-YOLO. Compared with YOLOV2 and the original Tiny-YOLO, the detection effect of the DF-Tiny-YOLO model was somewhat improved. The mAP of DF-Tiny-YOLO is 99.99%, which was 0.17% higher than that of Tiny-YOLO. The average IoU was 90.88%, which was 1.59% and 10.38% higher than YOLOV2 and Tiny-YOLO, respectively. Recall was 1.00, which was the same as YOLOV2 and 0.01 higher than Tiny-YOLO. The detection speed was 280 FPS, which was the same as that of Tiny-YOLO, and the number of images that can be processed per second was 3-fold that of YOLOv2. Standard deviations and confidence intervals for each indicator are also shown here.
https://doi.org/10.1371/journal.pone.0262629.t005
The confusion matrix is a standard format for representing accuracy evaluation and is represented in this paper in the form of a matrix with 4 rows and 4 columns. In this experiment, the total number of each row of the confusion matrix represents the number of leaves predicted to be in that category, while each column represents the true attribution category of apple leaves. Table 6 shows the confusion matrix of DF-Tiny-YOLO, including the classification precision and Recall of four different classes of diseases. The matrix results reveal that the model mAP is 99.99% and the average IoU is 90.88%, indicating that the majority of the prediction results are consistent with the true situation and the model performs best for Apple cedar rust and healthy leaves.
https://doi.org/10.1371/journal.pone.0262629.t006
Fig 7 shows the detection of apple leaf diseases by DF-Tiny-YOLO, YOLOv2, and Tiny-YOLO. Based on output image results, each network model might have a specific degree of deviation, but the positioning box of DF-Tiny-YOLO was more in line with the target sample, the results were more inclusive, and the error rate was lower. This further showed that the proposed network model has specific advantages.
The figures show the classification and localization effects of different models.
https://doi.org/10.1371/journal.pone.0262629.g007
4. Conclusion
4.1 research conclusion.
Nowadays, much progress has been made in the detection of apple leaf diseases. When the traditional approach seemed to have hit a bottleneck, higher accuracy can be realized more easily using a deep learning approach based on convolutional neural networks. Especially, since target detection technology appeared during the 1960s, target detection based on deep learning has emerged, and convolutional neural network learning methods are now being applied to classify and identify apple leaf diseases, such as the SDD model, and the R-CNN and YOLO series of algorithms. Many improved schemes have been proposed based on existing algorithms.
Redmon, Divvala [ 38 ] launched the YOLOv1 target detection network in 2016 and it has since been improved in versions v2 and v3. The Tiny-YOLO model reduces the model parameter setting and accelerates the model training process. The present study improved the Tiny-YOLO model by combining DenseNet and F-YOLO network concepts and we propose that the DF-Tiny-YOLO apple leaf disease detection model can solve the problem of differentiating apple scab, black rot, and cedar rust from healthy apple leaves. These diseases are complicated, difficult to identify and have a low recognition rate among similar diseases. Our experimental results showed that the mAP of DF-Tiny-YOLO was 99.99%, the average IoU was 90.88%, and the detection speed was 280 FPS. Compared with previous studies done by scholars for apple leaf disease identification [ 29 – 32 ], DF-Tiny-YOLO has three major innovation: Firstly, this paper proposed a more advanced method for identifying the complex texture of apple leaves and differentiating similar diseases, which helps growers to accurately judge and rapidly deal with diseases, which further benefits growers. Secondly, in terms of the model structure, based on the model, This study optimized the structure of the original YOLO convolutional neural network model at three levels, and proposed the new DF-Tiny-YOLO network model, which detected apple leaf disease faster and more accurately. Thirdly, the results of comparative experimental exploration showed that among YOLOv2, Tiny-YOLO, the optimized DF-Tiny-YOLO network offered more advantages than the other two models. Therefore, we believe that the DF-Tiny-YOLO model can detect apple leaf diseases quickly and effectively.
Upgraded, more complex versions of the YOLO network algorithm have been developed. However, we selected Tiny-YOLO as the basic network for improvement because the model is simplified and fewer parameters need to be trained. Together with the combination of dense network and F-YOLO, the network model has further acquired more advantages based on maintaining its own advantages, which is an interesting innovation. The present study on the one hand, proved the effectiveness of our experimental enhancements and provided some experience with the development of current methods of apple leaf disease detection. On the other hand, more effective experiments can be conducted on this basis in the future.
4.2 Future research
Due to time, environmental and other constraints, there are still several areas for improvement and further expansion of the research in this paper.
- Data availability is a prerequisite for research, and in apple leaf disease detection research, sufficient and diverse raw training data can help the effective training of detection models. At present, there are limited image data available on the Internet, and cooperation with apple plantations and agricultural research institutions is a useful and effective option.
- Various new convolutional neural network models are emerging and existing models are being improved and developed, and more target detection models suitable for apple leaf disease detection are yet to be explored. However, the study is limited by the experimental environment, which makes it difficult to compare with more models. The future research work will focus on other excellent detection models and gradually optimize the results of apple leaf disease detection by using appropriate data enhancement techniques and network structure improvement methods.
- In addition to apple leaf disease images, videos can also be used as detection objects, and real-time detection of more types of apple leaves during video shooting, etc. will also be the next step for effective implementation in subsequent research. With the deep learning technology in the field of target detection, several detection techniques have been effectively integrated, more and more new theories and methods have been proposed, and the accuracy and real-time performance of target detection have been improved, which will further help the research of apple leaf disease detection based on convolutional neural network.
Supporting information
S1 file. experimental results..
The file shows detailed results of experiments.
https://doi.org/10.1371/journal.pone.0262629.s001
https://doi.org/10.1371/journal.pone.0262629.s002
- View Article
- Google Scholar
- PubMed/NCBI
- 13. Krizhevsky A, Sutskever I, Hinton G, editors. ImageNet Classification with Deep Convolutional Neural Networks. NIPS; 2012.
- 37. Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu CY, et al., editors. SSD: Single Shot MultiBox Detector. European Conference on Computer Vision; 2016.
- 39. Redmon J, Farhadi A, editors. YOLO9000: Better, Faster, Stronger. IEEE Conference on Computer Vision & Pattern Recognition; 2017.
- DOI: 10.1016/J.MICPRO.2021.104027
- Corpus ID: 234317550
Recognition of Apple Leaf Diseases using Deep Learning and Variances-Controlled Features Reduction
- Muqadas Bin Tahir , M. A. Khan , +4 authors Muhammad Nazir
- Published 15 January 2021
- Agricultural and Food Sciences, Computer Science
- Microprocessors and Microsystems
42 Citations
Deep convolutional neural network-based framework for apple leaves disease detection, detection of apple plant diseases using leaf images through convolutional neural network, citrus diseases recognition using deep improved genetic algorithm, a cascaded design of best features selection for fruit diseases recognition, early-stage apple leaf disease prediction using deep learning, improved deep residual network forapple leaf disease identification, classification of citrus plant diseases using deep transfer learning, automatic crop disease recognition by improved abnormality segmentation along with heuristic-based concatenated deep learning model, cucumber leaf diseases recognition using multi level deep entropy-elm feature selection, cotton leaf diseases recognition using deep learning and genetic algorithm, 61 references, fruits diseases classification: exploiting a hierarchical framework for deep features fusion and selection, entropy‐controlled deep features selection framework for grape leaf diseases recognition, deep learning in plant diseases detection for agricultural crops: a survey, banana leaf diseased image classification using novel heap auto encoder (hae) deep learning, ccdf: automatic system for segmentation and recognition of fruit crops diseases based on correlation coefficient and deep cnn features, research on deep learning in apple leaf disease recognition, identification of apple leaf diseases based on deep convolutional neural networks, automatic image-based plant disease severity estimation using deep learning, using deep learning for image-based plant disease detection, a citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning, related papers.
Showing 1 through 3 of 0 Related Papers
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

A method of detecting apple leaf diseases based on improved convolutional neural network
Center for Innovation Management Research of Xinjiang, School of Economics and Management, Xinjiang University, Urumqi, Xinjiang, China
Associated Data
All data files are available from the PlantVillage database ( https://plantvillage.org/ ). These are third party data (i.e., data not owned or collected by the author(s)) and we confirm that we did not have any special access privileges that others would not have.
Apple tree diseases have perplexed orchard farmers for several years. At present, numerous studies have investigated deep learning for fruit and vegetable crop disease detection. Because of the complexity and variety of apple leaf veins and the difficulty in judging similar diseases, a new target detection model of apple leaf diseases DF-Tiny-YOLO, based on deep learning, is proposed to realize faster and more effective automatic detection of apple leaf diseases. Four common apple leaf diseases, including 1,404 images, were selected for data modeling and method evaluation, and made three main improvements. Feature reuse was combined with the DenseNet densely connected network and further realized to reduce the disappearance of the deep gradient, thus strengthening feature propagation and improving detection accuracy. We introduced Resize and Re-organization (Reorg) and conducted convolution kernel compression to reduce the calculation parameters of the model, improve the operating detection speed, and allow feature stacking to achieve feature fusion. The network terminal uses convolution kernels of 1 × 1, 1 × 1, and 3 × 3, in turn, to realize the dimensionality reduction of features and increase network depth without increasing computational complexity, thus further improving the detection accuracy. The results showed that the mean average precision (mAP) and average intersection over union (IoU) of the DF-Tiny-YOLO model were 99.99% and 90.88%, respectively, and the detection speed reached 280 FPS. Compared with the Tiny-YOLO and YOLOv2 network models, the new method proposed in this paper significantly improves the detection performance. It can also detect apple leaf diseases quickly and effectively.
1. Introduction
With over 2,000 years of history in China, areas of apple implantation have expanded annually, and 65% of all apples in the world are now produced in China. The traditional apple industry has been modernized with the support of national economic policies, and China has gradually developed into a major power source in the apple industry. Nonetheless, behind the rapid development of the apple implantation industry, disease prevention and control have been important problems that have long perplexed orchard farmers [ 1 ], who identify apple diseases by referring to experience, books, the Internet, and consulting professional and technical personnel [ 2 ]. However, relying solely on these sources is not conducive to timely and effective identification of diseases, and might even cause other problems associated with subjective judgment. Effective automated detection of apple diseases during production not only promptly monitors the health status of apples but also helps orchard farmers to correctly judge apple diseases. They can then implement timely prevention and control to avoid large-scale diseases, which is crucial for promoting the healthy growth of apples and increasing the economic benefits of orchards [ 3 , 4 ].
Hundreds of apple diseases disseminate in fruits, leaves, branches, roots, and other areas, but often initially appear in leaves, which are easily observed, collected, and managed. Therefore, they are an important reference for disease identification and effective automated detection of diseases is essential. However, judging differences among diseases is difficult due to the complexity of blade veins [ 5 ], resulting in unsatisfactory outcomes of experimental detection methods [ 6 ].
Nowadays, based on traditional algorithms, much methods has been made in the detection of apple leaf diseases, artificial neural network algorithms, genetic algorithms [ 7 ], support vector machines (SVMs), and a clustering algorithm [ 8 ] have applied feature extraction to apple leaves, and Li and He [ 9 ] established a BP network model with intelligent recognition that can diagnose apple leaf diseases using low-resolution images. Rong-Ming, Wei [ 10 ] proposed a method for extracting apple leaf disease spots based on hyperspectral images. Omrani, Khoshnevisan [ 11 ] proposed a calculation based on an artificial neural network (ANN) and SVMs to detect three apple leaf diseases. Shuaibu, Lee [ 12 ] applied hyperspectral images to detect marssonina blotch in apples at different stages and used orthogonal subspace projection for feature selection and redundancy reduction which generated better results. Zhang, Zhang [ 7 ] used genetic algorithm and feature selection method based on correlation to identify three kinds of apple leaf diseases, with an average recognition rate of 90%. Although traditional methods can also extract a large number of classification features, to select a few optimal features using general feature selection methods is difficult, so the recognition rate of these methods is low and generalization ability is weak.
Since the emergence of target detection technology during the 1960s, many domestic and overseas investigators have focused on the development of new technologies. After Krizhevsky, Sutskever [ 13 ] proposed the AlexNet structure at the ImageNet Large-scale Visual Recognition Challenge (ILSVRC) in 2012, target detection based on deep learning has gradually become mainstream [ 14 ]. Instead of using manual feature extraction, deep learning adopts hierarchical feature extraction [ 15 ], which is more similar to the human visual mechanism and has more accurate detection [ 16 ]. Many investigations into target detection based on deep learning have emerged [ 17 ], and applications to fruit and vegetable crop disease detection have been continuously proposed. Based on the AlexNet model, Brahimi, Boukhalfa [ 18 ] classified nine tomato diseases using more than 10,000 tomato disease images with a simple background in the PlantVillage database, further enhancing the model recognition rate. Priyadharshini, Arivazhagan [ 19 ] proposed a deep CNN-based architecture (modified LeNet) for four maize leaf disease classifications, and the trained model achieved 97.89% accuracy. In order to effectively examine the minor disease spots on diseased grape leaves, Zhu, Cheng [ 20 ] proposed a detection algorithm based on convolutional neural network and super-resolution image enhancement, the test effect is greatly improved. Zhang, Zhang [ 21 ] improved the model structure of the LeNet convolutional neural network to solve the problems of leaf shape, illumination, and image background that easily affect detection using traditional recognition methods; their model recognized six cucumber diseases, with an average recognition ratio of 90.32%. Xue, Huang [ 22 ], overcame the difficulty of detecting mango overlap or occlusion and proposed the improved Tiny-YOLO mango detection network combining dense connections, with an accuracy rate of 97.02%. Xiao, Li-ren [ 23 ] proposed the feature pyramid networks-single-shot multibox detector (FPN-SSD) model to detect top soybean leaflets on collected images, and automatically detect soybean leaves and classify leaf morphology. The wide application of deep learning to the detection of fruit and vegetable diseases has furthered automatic disease detection by leaps and bounds [ 24 , 25 ].
Convolutional neural network learning methods are now being applied to classify and identify apple leaf diseases, such as the SDD model, and the R-CNN and YOLO series of algorithms. Many improved schemes have been proposed based on existing algorithms. Jiang, Chen [ 26 ] proposed the INAR-SSD (SSD with an Inception module and Rainbow concatenation) model to detect five common apple leaf diseases, with high accuracy (78.80% mAP) in real time. The INAR-SSD has provided feasible real-time detection of apple leaf diseases. Liu, Zhang [ 27 ] created a new structure based on the deep convolutional neural network AlexNet to detect apple leaf diseases, thus further improving the effective identification of apple leaf diseases. Based on the DenseNet-121 deep convolutional network, Zhong and Zhao [ 28 ] proposed regression, multi-label classification, and focus loss function, to identify apple leaf diseases, with a recognition accuracy rate > 93%, which was better than the traditional multi-classification method based on the cross-entropy loss function. Jan and Ahmad [ 29 ] developed an apple pest and disease diagnostic system to predict apple scabs and leaf spots. Entropy, energy, inverse difference moment (IDM), mean, standard deviation (SD), and perimeter, etc., were extracted from apple leaf images and a multi-layer perceptron (MLP) pattern classifier and 11 apple leaf image features to train the model, which had 99.1% diagnostic accuracy. Yu, Son [ 30 ] proposed a new method based on a region-of-interest-aware deep convolutional neural network (ROI-aware DCNN) to render deep features more discriminative and increase classification performance for apple leaf disease identification, with an average accuracy of 84.3%. Sun, Xu [ 31 ] proposed a lightweight CNN model that can be deployed on mobile devices to detect apple leaf diseases in real time to test five common apple leaf diseases, with a mAP value of 83.12%. Di and Qu [ 32 ] applied the Tiny-YOLO model to apple leaf diseases, and the results showed that the model was effective.
There are two main types of target detection in the deep learning era [ 33 ]. The first is based on region formation. Representative models are R-CNN [ 34 ], Fast R-CNN [ 35 ], and Faster R-CNN [ 36 ]. One method forms candidate boxes that might contain target areas in images, extracts features from each box, trains classifiers, and achieves the target detection of specific objects in two steps. The other method is based on regression. Typical representative models are SDD [ 37 ], YOLO [ 38 ], and YOLOv2 [ 39 ]. These methods address detection as regression problems; they directly predict the coordinates of the boxes, the confidence level of the objects contained, and the probability of the objects appearing, then realizes the target location and identifies the category of specific objects in one step. The detection accuracy of the region formation-based method is superior. However, a target detection method based on regression is more advantageous in terms of detection speed and is popular in terms of real-time detection and intelligent applications. Yet, most recent research efforts focusing on detecting apple leaf diseases are not enough, and considerable room exists for improving detection accuracy and effectiveness. Therefore, the present study investigated how to optimize methods of apple leaf disease detection. Practical problems such as complexity and variations in the veins of apple leaves and difficulties with disease identification can be overcome by the rapid and effective automated detection of apple leaf diseases. Therefore, we created the DF-Tiny-YOLO model to, an detect apple leaf diseases based on regression with the concepts of DenseNet [ 40 ] and F-YOLO [ 41 ]. We then optimized the model considering rapid, accurate detection.
2. Algorithm principle and DF-Tiny-YOLO network
2.1 algorithm principles, 2.1.1 yolo network model.
The core principle of YOLO is the consideration that target detection is a regression problem. In this algorithm, images are divided into S × S grid cells, where each grid predicts B bounding boxes, and each bounding box contains five predictive values ( x , y , w , h , confidence ): x , y indicates the central coordinate of the prediction bounding box relative to the grid boundary, and w , h indicates the width and height of the prediction bounding box. Confidence has two definitions: one is the possibility that a bounding box contains a target, and the other is the prediction accuracy of the bounding box, expressed as the IoU of the joint between the prediction box and the actual bounding box, defined as:
In Eq (1) , Pr ( object ) is the probability that a target object falls into the grid, which is a standard for measuring the detection accuracy of a specific object in a dataset. If a target object is not in the grid, the Pr ( object ) value is 0, and the final value is 0; otherwise, the Pr ( object ) value is 1, at this point the final confidence value is I o U p r e d i c t i o n t r u t h . The conditional class probability of each grid ( Pr ( Class i | object )) is multiplied by the confidence to obtain the confidence of the specific class of objects contained in the prediction box Eq (2) :
On this basis, YOLO selects the appropriate candidate region to predict while detecting the input picture to obtain the optimal position and classification of objects in the candidate region. The YOLO network model comprises 24 convolutions and two full-link layers. Redmon, Divvala [ 38 ] applied the YOLO algorithm to various datasets to verify its accuracy. The average YOLO target detection accuracy is about 60%, with a recognition rate up to 45 frames/s.
2.1.2 YOLOv2 network
Although YOLO has been greatly improved in terms of detection speed, its detection accuracy is far from optimal. Object positioning is insufficiently accurate, and its recall rate is relatively low. Thus, YOLOv2 has been improved based on the advantages of the original YOLO, which proposes the new network structure, Darenet-19, which includes 19 convolution and five maximum pooling layers and adopts a series of optimization strategies. In terms of candidate box selection, the K-means clustering method is adopted in YOLOv2 to determine the size, proportion, and number of candidate boxes, and obtain reasonable candidate box selection parameters through balanced clustering results and computational complexity. A pass-through layer has been added to the network structure to connect shallow and deep features, which has adapted YOLOv2 to multi-scale features and improved the detection accuracy of small targets. Multi-scale training was adopted, and a group of new pictures was randomly selected for scale input every 10 rounds of training to enhance detection performance. Moreover, many aspects have been improved. The final test results of the VOC 2007 dataset showed that when the size of the input picture was 228 × 228, the frame rate reached 67 FPS and mAP reached 76.8%. When the input picture resolution was 554 × 554, the mAP of YOLOv2 on VOC 2007 reached 78.6% and the frame rate reached 40 FPS, which satisfied real-time requirements.
2.1.3 Tiny-YOLO network
Tiny-YOLO is a miniature “accelerated version” network based on the Darknet-19 structure ( Fig 1 ). This reduces the complexity of the model, and the number of training parameters greatly improves the training speed up to 244 frames/s, which is 1.5-fold the detection speed of YOLOv2. However, noise increased during Tiny-YOLO training due to reduced model depth. Despite progress in speed, target positioning accordingly became inaccurate, resulting in a low average accuracy of 23.7%, compared with YOLOv2, which can achieve 78.6% accuracy at 40 frames/s. The detection accuracy of Tiny-YOLO can still be improved.

Tiny-YOLO structure and the network parameter shown in here.
2.1.4 Dense connective network
Huang, Liu [ 40 ] connected all layers in the network in pairs so that each layer in the network can receive the characteristics of all layers in front of it as input. This maximized information flow among all layers in the network. This network structure is called DenseNet because of its numerous dense connections ( Fig 2 ).

DenseNet has the following important characteristics. It alleviates the problem of gradient dissipation during training to some extent. Each layer receives gradient signals from all following layers during reverse transmission, and the gradient near the input layer will not become smaller as the network depth increases. As numerous features are reused, many features can be generated using fewer convolution kernels, so the size of the final model is relatively small. The main purpose of DenseNet was to establish a connection relationship between different layers. In the traditional convolution structure, layer k has k connections, whereas in the DenseNet structure, layer k has k ( k − 1)/2 connections, that is, each layer in the model is connected with the following layer to enhance feature extraction. Fig 3 shows the network structure of DenseNet. Overall, the structure consists mainly of dense blocks and transition layers. The internal feature diagrams of the dense blocks are of the same size, and transition block nodes comprise “BN-Conv-Pool”.

DenseNet encourages feature reuse, which means that the features of each picture can be used, and this reduces disappearance of feature gradient, strengthens the feature propagation of the network model, and effectively inhibits the overfitting problem.
2.1.5 F-YOLO network
Mehta and Ozturk [ 41 ] proposed F-YOLO based on Tiny-YOLO. Generally, a deeper and wider network structure, results in a better detection effect, but the corresponding calculation of parameters will also increase, resulting in slow network training. Considering this problem, Mehta et al. used the resize in the YOLOv2 algorithm to compress the convolution layers of the 6th, 8th, and 10th layers by a 1 × 1 convolution kernel, superimpose them on the first 13 × 13 × 1024 convolution layer, and connect them to form a 13 × 13 × 1792 feature diagram. Thereafter, many 1 × 1 convolution layers were added to increase the network depth without increasing the computational complexity, thus producing a “narrow and deep” network. Compared with the original Tiny-YOLO network, the modified network improved the accuracy by 5 mAP and the speed by 20%. On the Pascal dataset, the training parameters of the final model were reduced > 10-fold, and the training speed exceeded 200 FPS compared with the VGG-based target detection model.
2.2 DF-Tiny-YOLO network model
The following improvements to the original Tiny-YOLO network structure were made based on existing knowledge to realize the rapid identification and accurate location of apple leaf diseases.
Huang, Liu [ 40 ] proposed the concept of a dense connection network. They replaced the 13 × 13 × 512 convolution layer in the Tiny-YOLO structure with a dense module; the 13 × 13 × 128 convolution kernel was used for convolution to output 128 feature diagrams, so that the spliced output of the input and first layers is 13 × 13 × (512 + 128), the output set of the second layer is 13 × 13 × (640 + 128), and so on. The spliced output of the last layer is 13 × 13 × 1024, thus reducing gradient disappearance, and further realizing feature reuse, strengthening feature propagation, and improving detection accuracy.
Resize and reorg were adopted to compress large feature diagrams, reduce network computation, accelerate network training, and improve the speed of detection. Large and small feature diagrams were stacked on the last main convolution layer to achieve feature fusion.
Using 1 × 1, 1 × 1, and 3 × 3 convolution kernels at the network terminals will contribute to reducing the dimension of features and increasing the network depth without increasing computational complexity, thus further improving the detection accuracy.
A DF-Tiny-YOLO network model was constructed with the structure shown in Fig 3 based on these improvements, and the network parameter also shown in here.
3. Analysis of experimental process and results
3.1 experimental process, 3.1.1 experimental data.
We randomly selected four clear common apple leaf pictures of the same size in the PlantVillage database as the experimental dataset. The quality of this dataset was ensured by manual screening to avoid the problems of singleness, repeatability, and errors. The data included 300 images of leaves with apple scab, 300 with apple black rot, 275 with apple cedar rust, and 529 healthy leaves, total 1,404 images. Fig 4 describes the basic situation of four types of leaves. The images were randomly divided into training and test sets at a ratio of 80%:20%, the division of data set was shown in Table 1 .

The figure describes the basic situation of four types of leaves.
| Classes | Raw data set | Training data set | Test data set |
|---|---|---|---|
| 529 | 423 | 106 | |
| 300 | 240 | 60 | |
| 300 | 240 | 60 | |
| 275 | 220 | 55 | |
| 1404 | 1123 | 281 |
The table shows division of training and test sets.
Many common diseases affect apple leaves. Table 2 summarizes the main characteristics of diseased and healthy leaves with reference to relevant information.
| Disease type | Characteristics |
|---|---|
| Elliptical or oval, with blunt serrations on leaf edge. Both sides of young leaves are pilose, and front pilose falls off when mature. | |
| Almost round or radial disease spots. Leaves are initially covered with green-brown mildew that later turns black. Diseased spots can cover whole leaves, causing a scorched appearance. | |
| Disease spots are initially small and purple-black. These expand into round spots with a yellowish-brown middle with brown purple edges resembling frog eyes. | |
| Small, oily orange-red disease spots gradually expand and become round orange-yellow and edged in red. |
After uniformly labeling the four categories of apple leaf picture data, the leaf samples were manually labeled individually according to the following criteria: label box frames apple leaves in the image regardless of the stem, and the upper, lower, left, and right sides of the label box are close to the outer boundary of the leaves, to minimize errors emerging during sample labeling.
3.1.2 Environment and tools
Table 3 shows the server platform configuration and tools used herein.
| Configuration Item | Value |
|---|---|
| Intel Core i7–8700K @ 3.70 GHz | |
| Nvidia GeForce GTX 1080 Ti GPU | |
| 64GB RAM | |
| Intel SSDSCKKW256H6 | |
| Windows 7 Ultimate 64bit SP1 | |
| TensorFlow 1.3+ |
3.1.3 Model training
Referring to the training parameters of the original Tiny-YOLO model, the main initial training parameters of the improved DF-Tiny-YOLO model set in this study are shown in the Table 4 . Where the learning rate parameter values will be adjusted appropriately during the model training period based on the average loss values.
| Parameter name | Parameter value |
|---|---|
| 64 | |
| 4 | |
| 0.9 | |
| 0.0005 | |
| 0.001 |
Fig 5 shows the experimental process for the target detection of apple leaf diseases. The apple leaf images are first screened, then the entire leaf data set is divided into a training set and a test set, and finally an experimental file containing the corresponding information is generated by annotating the samples. Using the apple leaf training data, the detection training is performed on the original YOLOv2 model, the Tiny-YOLO model, and the modified DF-Tiny-YOLO model, respectively. The DF-Tiny-YOLO network model was generated by improving the Tiny-YOLO network through combining DenseNet with the F-YOLO concept. The model was trained and tested using the training and test datasets, respectively, since the measurement error decreases in many iterations, the network performance is optimized with the correction of parameters. The training ends when the average loss value of the network model decreases to a certain level and the value does not decrease in subsequent iterations, to generate the DF-Tiny-YOLO apple leaf disease detection model. The experimental findings of DF-Tiny-YOLO, YOLOv2, and the original Tiny-YOLO were compared.

The figure shows the experimental process for the target detection of apple leaf diseases.
3.1.4 Experimental evaluation indexes
The mean average precision (mAP), intersection over union (IoU), recall, and frame per second (FPS) were adopted as the evaluation indexes of the model to detect apple leaf diseases. The results were assessed as true positive (TP), true negative (TN), false positive (FP), and false negative (FN).
Precision (accuracy rate), is the proportion of correctly recognized object A to the number of all objects recognized as A in the recognition result. Mean average precision is the result of averaging the accuracy rates of all types ( Eq (3) ).
Recall (recall ratio), indicates the proportion of the number of objects correctly identified as A in the recognition result to the number of all objects identified as A in the test set ( Eq (4) )
The IoU refers to the degree of overlap between the prediction box of object A and the original label box ( Eq (5) ).
Frame Rate per Second (FPS) is a common indicator of speed, namely, the number of pictures that can be processed per second.
3.2 Experimental results and analysis
To prevent the prediction model from overfitting, we determine whether the training model is optimal by observing the average loss value. We end the training when the average loss value decreases to a certain level and the value does not decrease again in the subsequent iterations. The training is stopped when the average loss of the network model decreases to the level of 0.0048. Fig 6 shows variations in the average losses of DF-Tiny-YOLO and Tiny-YOLO during training. Generally, the model rapidly fits at the start of the iteration, and the loss also rapidly decreases. With the continuous increase in the number of iterations, the loss gradually tended to stabilize during forward and backward oscillation. The loss values of DF-Tiny-YOLO and Tiny-YOLO in the first iteration were 23.79% and 28.46%, respectively. As the number of iterations increased, the loss values decreased faster in DF-Tiny-YOLO than Tiny-YOLO. The model fitting was faster, the oscillation amplitude is more moderate, and the final stationary value was relatively smaller. These results show that the proposed DF-Tiny-YOLO model has better training effects.

The figure shows variations in the average losses of DF-Tiny-YOLO and Tiny-YOLO during training.
Table 5 compares the experimental results of DF-Tiny-YOLO, YOLOv2, and Tiny-YOLO. Compared with YOLOV2 and the original Tiny-YOLO, the detection effect of the DF-Tiny-YOLO model was somewhat improved. The mAP of DF-Tiny-YOLO is 99.99%, which was 0.17% higher than that of Tiny-YOLO. The average IoU was 90.88%, which was 1.59% and 10.38% higher than YOLOV2 and Tiny-YOLO, respectively. Recall was 1.00, which was the same as YOLOV2 and 0.01 higher than Tiny-YOLO. The detection speed was 280 FPS, which was the same as that of Tiny-YOLO, and the number of images that can be processed per second was 3-fold that of YOLOv2. Standard deviations and confidence intervals for each indicator are also shown here.
| Training Network | Variable | Mean | Std. Err. | 95% Conf. Interval |
|---|---|---|---|---|
| mAP | 99.99% | 0.01 | [99.97, 100.00] | |
| Average IoU | 90.88% | 0.12 | [90.62, 91.14] | |
| Recall | 1.00 | 0.00 | [1.00, 1.00] | |
| FPS | 280 | |||
| mAP | 99.99% | 0.01 | [99.98, 100.00] | |
| Average IoU | 89.29% | 0.49 | [88.19, 90.39] | |
| Recall | 1.00 | 0.00 | [0.99, 1.00] | |
| FPS | 93 | |||
| mAP | 99.81% | 0.03 | [99.74, 99.88] | |
| Average IoU | 80.50% | 1.78 | [76.47, 84.53] | |
| Recall | 0.99 | 0.00 | [0.98, 0.99] | |
| FPS | 280 | |||
The confusion matrix is a standard format for representing accuracy evaluation and is represented in this paper in the form of a matrix with 4 rows and 4 columns. In this experiment, the total number of each row of the confusion matrix represents the number of leaves predicted to be in that category, while each column represents the true attribution category of apple leaves. Table 6 shows the confusion matrix of DF-Tiny-YOLO, including the classification precision and Recall of four different classes of diseases. The matrix results reveal that the model mAP is 99.99% and the average IoU is 90.88%, indicating that the majority of the prediction results are consistent with the true situation and the model performs best for Apple cedar rust and healthy leaves.
| Classes | Apple Scab | Black Rot | Cedar Apple Rust | Healthy | Precision% | Recall |
|---|---|---|---|---|---|---|
| 299 | 1 | 0 | 0 | 99.99 | 1.00 | |
| 1 | 299 | 0 | 0 | 99.97 | 1.00 | |
| 0 | 0 | 275 | 0 | 100.00 | 1.00 | |
| 0 | 0 | 0 | 529 | 100.00 | 1.00 | |
| 99.99% | ||||||
| 90.88% | ||||||
The table shows the confusion matrix of DF-Tiny-YOLO, including the classification precision and Recall of four different classes of diseases.
Fig 7 shows the detection of apple leaf diseases by DF-Tiny-YOLO, YOLOv2, and Tiny-YOLO. Based on output image results, each network model might have a specific degree of deviation, but the positioning box of DF-Tiny-YOLO was more in line with the target sample, the results were more inclusive, and the error rate was lower. This further showed that the proposed network model has specific advantages.

The figures show the classification and localization effects of different models.
4. Conclusion
4.1 research conclusion.
Nowadays, much progress has been made in the detection of apple leaf diseases. When the traditional approach seemed to have hit a bottleneck, higher accuracy can be realized more easily using a deep learning approach based on convolutional neural networks. Especially, since target detection technology appeared during the 1960s, target detection based on deep learning has emerged, and convolutional neural network learning methods are now being applied to classify and identify apple leaf diseases, such as the SDD model, and the R-CNN and YOLO series of algorithms. Many improved schemes have been proposed based on existing algorithms.
Redmon, Divvala [ 38 ] launched the YOLOv1 target detection network in 2016 and it has since been improved in versions v2 and v3. The Tiny-YOLO model reduces the model parameter setting and accelerates the model training process. The present study improved the Tiny-YOLO model by combining DenseNet and F-YOLO network concepts and we propose that the DF-Tiny-YOLO apple leaf disease detection model can solve the problem of differentiating apple scab, black rot, and cedar rust from healthy apple leaves. These diseases are complicated, difficult to identify and have a low recognition rate among similar diseases. Our experimental results showed that the mAP of DF-Tiny-YOLO was 99.99%, the average IoU was 90.88%, and the detection speed was 280 FPS. Compared with previous studies done by scholars for apple leaf disease identification [ 29 – 32 ], DF-Tiny-YOLO has three major innovation: Firstly, this paper proposed a more advanced method for identifying the complex texture of apple leaves and differentiating similar diseases, which helps growers to accurately judge and rapidly deal with diseases, which further benefits growers. Secondly, in terms of the model structure, based on the model, This study optimized the structure of the original YOLO convolutional neural network model at three levels, and proposed the new DF-Tiny-YOLO network model, which detected apple leaf disease faster and more accurately. Thirdly, the results of comparative experimental exploration showed that among YOLOv2, Tiny-YOLO, the optimized DF-Tiny-YOLO network offered more advantages than the other two models. Therefore, we believe that the DF-Tiny-YOLO model can detect apple leaf diseases quickly and effectively.
Upgraded, more complex versions of the YOLO network algorithm have been developed. However, we selected Tiny-YOLO as the basic network for improvement because the model is simplified and fewer parameters need to be trained. Together with the combination of dense network and F-YOLO, the network model has further acquired more advantages based on maintaining its own advantages, which is an interesting innovation. The present study on the one hand, proved the effectiveness of our experimental enhancements and provided some experience with the development of current methods of apple leaf disease detection. On the other hand, more effective experiments can be conducted on this basis in the future.
4.2 Future research
Due to time, environmental and other constraints, there are still several areas for improvement and further expansion of the research in this paper.
- Data availability is a prerequisite for research, and in apple leaf disease detection research, sufficient and diverse raw training data can help the effective training of detection models. At present, there are limited image data available on the Internet, and cooperation with apple plantations and agricultural research institutions is a useful and effective option.
- Various new convolutional neural network models are emerging and existing models are being improved and developed, and more target detection models suitable for apple leaf disease detection are yet to be explored. However, the study is limited by the experimental environment, which makes it difficult to compare with more models. The future research work will focus on other excellent detection models and gradually optimize the results of apple leaf disease detection by using appropriate data enhancement techniques and network structure improvement methods.
- In addition to apple leaf disease images, videos can also be used as detection objects, and real-time detection of more types of apple leaves during video shooting, etc. will also be the next step for effective implementation in subsequent research. With the deep learning technology in the field of target detection, several detection techniques have been effectively integrated, more and more new theories and methods have been proposed, and the accuracy and real-time performance of target detection have been improved, which will further help the research of apple leaf disease detection based on convolutional neural network.
Supporting information
The file shows detailed results of experiments.
Funding Statement
National Natural Science Foundation of China (82060265)” and “the Department of Human Resources and Social Security of Xinjiang Uygur Autonomous Region (042419001).
Data Availability
Classification of Apple Plant Leaf Diseases Using Deep Convolutional Neural Network
- CC BY-NC-ND 4.0
- This person is not on ResearchGate, or hasn't claimed this research yet.

- University of Gujrat

Abstract and Figures

Discover the world's research
- 25+ million members
- 160+ million publication pages
- 2.3+ billion citations
- Priya Ujawe

- Bingqian Xing

- Zhongzhu Cao

- CMC-COMPUT MATER CON
- Samra Rehman

- Majed Alhaisoni
- Byoungchol Chang

- Shujuan Zhang
- Shilan Hong
- Zhaohui Jiang

- Jianpeng Xu

- Priyanka Pradhan

- Shashank Mohan
- Shenbagam Palanisamy
- Arunadevi A
- Rajasimman K
- Recruit researchers
- Join for free
- Login Email Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google Welcome back! Please log in. Email · Hint Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google No account? Sign up
Detection of plant leaf disease using advanced deep learning architectures
- Original Research
- Published: 29 May 2024
Cite this article

- Rakhee Sharma 1 ,
- Mamta Mittal 2 ,
- Vedika Gupta 3 &
- Dipit Vasdev 4
48 Accesses
Explore all metrics
Food is the basic necessity for human survival and plant diseases act as a critical hindrance to the quality of harvested crops. Timely identification and administration of plant diseases are remarkably essential owing to the ever-increasing population and global climate change. Though many algorithms have been designed for early diagnosis of plant leaf diseases in existing literature, the bulk of those lack a large enough dataset for accurate detection and diagnosis. This work aimed to determine the significance and perform fine-tuning of state-of-the-art models to detect and classify diseases in plant leaves. These models are required to perform early detection of diseases to prevent the loss of crops. In this paper, a dataset containing 39 classes of diseased and healthy leaves of 14 plants is used. We perform fine-tuning of various deep learning architectures including VGG16, AlexNet, ResNet18, and MobileNetV2. The results were evaluated based on four different metrics vis-à-vis accuracy, F1-score, precision, and recall. The best results were received using MobileNetV2 with an accuracy of 94.4%.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions

Source: Geetharamani, G. et al. (2019)

Similar content being viewed by others

Plant Leaf Diseases Detection Using Deep Learning Algorithms

Enhancing the performance of transferred efficientnet models in leaf image-based plant disease classification

Detection and Classification of Fruit Tree Leaf Disease Using Deep Learning
Stephen A, Arumugam P, Arumugam C (2024) An efficient deep learning with a big data-based cotton plant monitoring system. Int J Inf tecnol 16(1):145–151. https://doi.org/10.1007/s41870-023-01536-9
Article Google Scholar
Green JL, Capizzi J, A systematic approach to diagnosing plant damage
Kumar Y, Koul A, Singla R, Ijaz MF (2023) Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda. J Ambient Intell Humaniz Comput 14(7):8459–8486. https://doi.org/10.1007/s12652-021-03612-z
Kusuma S, Jothi KR (2024) Early betel leaf disease detection using vision transformer and deep learning algorithms. Int J Inf Tecnol 16(1):169–180. https://doi.org/10.1007/s41870-023-01647-3
Gupta V et al (2022) Improved COVID-19 detection with chest x-ray images using deep learning. Multimed Tools Appl 81(26):37657–37680. https://doi.org/10.1007/s11042-022-13509-4
Sachdeva J, Gupta M, Gupta S, Aggarwal D, Gupta V (2021) Review of signboard transliteration using deep learning. In: Balas VE, Hassanien AE, Chakrabarti S, Mandal L (eds) Proceedings of international conference on computational intelligence, in lecture notes on data science and cloud computing, data engineering and communications technologies. Springer, Singapore, pp 407–418. https://doi.org/10.1007/978-981-33-4968-1_32
Chapter Google Scholar
Geetharamani G, Pandian JA (2019) Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput Electr Eng 76:323–338. https://doi.org/10.1016/j.compeleceng.2019.04.011
Ashwinkumar S, Rajagopal S, Manimaran V, Jegajothi B (2022) Automated plant leaf disease detection and classification using optimal MobileNet based convolutional neural networks. Mater Today Proc 51:480–487. https://doi.org/10.1016/j.matpr.2021.05.584
Nazeer R et al (2024) Detection of cotton leaf curl disease’s susceptibility scale level based on deep learning. J Cloud Comp 13(1):50. https://doi.org/10.1186/s13677-023-00582-9
Tang Z et al (2023) A precise image-based tomato leaf disease detection approach using PLPNet. Plant Phenomics 5:0042. https://doi.org/10.34133/plantphenomics.0042
Kothari D, Mishra H, Gharat M, Pandey V, Thakur R (2022) Potato leaf disease detection using deep learning. Int J Eng Techn Res. https://doi.org/10.17577/IJERTV11IS110029
Azim M, Islam M, Rahman M, Jahan F (2021) An effective feature extraction method for rice leaf disease classification. TELKOMNIKA (Telecommun Comput Electron Contr) 19:463–470. https://doi.org/10.12928/telkomnika.v19i2.16488
Kaur N (2021) Plant leaf disease detection using ensemble classification and feature extraction. Turk J Comput Math Educ (TURCOMAT) 12(11):2339–2352
Google Scholar
Plants | Free Full-Text | Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data. Accessed: Dec. 20, 2023. [Online]. https://www.mdpi.com/2223-7747/10/1/28
Ganatra N, Patel A (2020) Performance analysis of fine-tuned convolutional neural network models for plant disease classification. Int J Control Autom 13(03):03
Jadhav SB, Udupi VR, Patil SB (2021) Identification of plant diseases using convolutional neural networks. Int J Inf Tecnol 13(6):2461–2470. https://doi.org/10.1007/s41870-020-00437-5
Khamparia A, Saini G, Gupta D, Khanna A, Tiwari S, de Albuquerque VHC (2020) Seasonal crops disease prediction and classification using deep convolutional encoder network. Circuits Syst Signal Process 39(2):818–836. https://doi.org/10.1007/s00034-019-01041-0
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran YA (2020) Using deep transfer learning for image-based plant disease identification. Comput Electron Agric 173:105393. https://doi.org/10.1016/j.compag.2020.105393
(PDF) Deep Learning-Based Techniques for Plant Diseases Recognition in Real-Field Scenarios. Accessed: Dec. 20, 2023. [Online]. Available: https://www.researchgate.net/publication/339062055_Deep_Learning-Based_Techniques_for_Plant_Diseases_Recognition_in_Real-Field_Scenarios
Sagar A, Jacob D (2021) On using transfer learning for plant disease detection. bioRxiv, p. 2020.05.22.110957. https://doi.org/10.1101/2020.05.22.110957
How Convolutional Neural Networks Diagnose Plant Disease | Plant Phenomics. Accessed: Dec. 20, 2023. [Online]. Available: https://spj.science.org/doi/ https://doi.org/10.34133/2019/9237136
Baranwal S, Khandelwal S, Arora A (2019) Deep learning convolutional neural network for apple leaves disease detection. Rochester, NY, Feb. 21. https://doi.org/10.2139/ssrn.3351641 .
Bharali P, Bhuyan C, Boruah A (2019) Plant disease detection by leaf image classification using convolutional neural network, 1025, 194–205, https://doi.org/10.1007/978-981-15-1384-8_16 .
Adedoja A, Owolawi PA, Mapayi T (2019) Deep learning based on NASNet for plant disease recognition using leave images, In: 2019 international conference on advances in big data, computing and data communication systems (icABCD), pp 1–5. https://doi.org/10.1109/ICABCD.2019.8851029
Francis M, Deisy C (2019) Disease detection and classification in agricultural plants using convolutional neural networks—a visual understanding, In: 2019 6th international conference on signal processing and integrated networks (SPIN), pp1063–1068. https://doi.org/10.1109/SPIN.2019.8711701
Too EC, Yujian L, Njuki S, Yingchun L (2019) A comparative study of fine-tuning deep learning models for plant disease identification. Comput Electron Agric 161:272–279. https://doi.org/10.1016/j.compag.2018.03.032
Türkoğlu M, Hanbay D (2019) Plant disease and pest detection using deep learning-based features. Turk J Electr Eng Comput Sci 27(3):1636–1651. https://doi.org/10.3906/elk-1809-181
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318. https://doi.org/10.1016/j.compag.2018.01.009
Gandhi R, Nimbalkar S, Yelamanchili N, Ponkshe S (2018) Plant disease detection using CNNs and GANs as an augmentative approach, In: 2018 IEEE international conference on innovative research and development (ICIRD), pp 1–5. https://doi.org/10.1109/ICIRD.2018.8376321
Deep Learning for Image Processing Applications, IOS Press. Accessed: Dec. 20, 2023. [Online]. https://www.iospress.com/catalog/books/deep-learning-for-image-processing-applications-0
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01419
Rakhee A, Singh A, Kumar A (2018) Weather based fuzzy regression models for prediction of rice yield. J Agrometeorol. https://doi.org/10.54386/jam.v20i4.569
Mittal M, Goyal LM, Kaur S, Kaur I, Verma A, Jude Hemanth D (2019) Deep learning based enhanced tumor segmentation approach for MR brain images. Appl Soft Comput 78:346–354. https://doi.org/10.1016/j.asoc.2019.02.036
Lundervold AS, Lundervold A (2019) An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 29(2):102–127. https://doi.org/10.1016/j.zemedi.2018.11.002
Jain N, Gupta V, Shubham S, Madan A, Chaudhary A, Santosh KC (2022) Understanding cartoon emotion using integrated deep neural network on large dataset. Neural Comput Appl 34(24):21481–21501. https://doi.org/10.1007/s00521-021-06003-9
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. arXiv, https://doi.org/10.48550/arXiv.1409.1556
Admass WS, Munaye YY, Bogale GA (2024) Convolutional neural networks and histogram-oriented gradients: a hybrid approach for automatic mango disease detection and classification. Int J Inf Tecnol 16(2):817–829. https://doi.org/10.1007/s41870-023-01605-z
Hemanth DJ, Anitha J, Balas VE (2015) Performance improved hybrid intelligent system for medical image classification, In: Proceedings of the 7th Balkan conference on informatics conference, in BCI ’15. New York, NY, USA: Association for Computing Machinery, pp 1–5. https://doi.org/10.1145/2801081.2801095
Shubham S, Jain N, Gupta V, Mohan S, Ariffin MM, Ahmadian A (2021) Identify glomeruli in human kidney tissue images using a deep learning approach. Soft Comput 27(5):2705–2716. https://doi.org/10.1007/s00500-021-06143-z
Gupta V, Gaur H, Vashishtha S, Das U, Singh VK, Hemanth DJ (2023) A fuzzy rule-based system with decision tree for breast cancer detection. IET Image Proc 17(7):2083–2096. https://doi.org/10.1049/ipr2.12774
Hemanth JD, Anitha J, Ane BK (2017) Fusion of artificial neural networks for learning capability enhancement: application to medical image classification. Expert Syst 34(6):e12225. https://doi.org/10.1111/exsy.12225
Krizhevsky A, Sutskever I, Hinton GE (2023) ImageNet classification with deep convolutional neural networks, In: Advances in neural information processing systems, curran associates, Inc., 2012. Accessed: Dec. 20, 2023. [Online]. https://proceedings.neurips.cc/paper_files/paper/2012/hash/c399862d3b9d6b76c8436e924a68c45b-Abstract.html
Shelar N, Shinde S, Sawant S, Dhumal S, Fakir K (2022) Plant disease detection using Cnn. ITM Web Conf 44:03049. https://doi.org/10.1051/itmconf/20224403049
Sujatha R, Chatterjee JM, Jhanjhi N, Brohi SN (2021) Performance of deep learning vs machine learning in plant leaf disease detection. Microprocess Microsyst 80:103615. https://doi.org/10.1016/j.micpro.2020.103615
Moupojou E et al (2023) FieldPlant: a dataset of field plant images for plant disease detection and classification with deep learning. IEEE Access 11:35398–35410. https://doi.org/10.1109/ACCESS.2023.3263042
Hu M, Long S, Wang C, Wang Z (2024) Leaf disease detection using deep convolutional neural networks. J Phys: Conf Ser. https://doi.org/10.1088/1742-6596/2711/1/012020
Download references
Author information
Authors and affiliations.
Center for Research and Innovation in Interdisciplinary Studies, BVICAM, New Delhi, India
Rakhee Sharma
Delhi Skill & Entrepreneurship University, New Delhi, India
Mamta Mittal
Jindal Global Business School, O.P. Jindal Global University, Sonipat, 131001, Haryana, India
Vedika Gupta
Department of Electrical and Computer Engineering, Tandon School of Engineering, New York University, Brooklyn, New York, USA
Dipit Vasdev
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Mamta Mittal .
Ethics declarations
Conflict of interest.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Sharma, R., Mittal, M., Gupta, V. et al. Detection of plant leaf disease using advanced deep learning architectures. Int. j. inf. tecnol. (2024). https://doi.org/10.1007/s41870-024-01937-4
Download citation
Received : 09 February 2024
Accepted : 14 May 2024
Published : 29 May 2024
DOI : https://doi.org/10.1007/s41870-024-01937-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Convolutional neural network (CNN)
- Deep learning
- Multi-class classification
- Plant leaf disease
- Plant village dataset
- Transfer learning
- Find a journal
- Publish with us
- Track your research
IEEE Account
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.

IMAGES
VIDEO
COMMENTS
Therefore, it is of great significance to study the identification of apple leaf diseases. Based on DenseNet-121 deep convolution network, three methods of regression, multi-label classification and focus loss function were proposed to identify apple leaf diseases. In this paper, the apple leaf image data set, including 2462 images of six apple ...
The experimental results show that the method proposed in this study achieves a recognition accuracy of 98.92%, which is better than that of other existing deep learning methods and achieves competitive performance on apple leaf disease identification tasks, which provides a reference for the application of deep learning methods in crop disease ...
To utilize the pretrained model for identifying plant diseases, this paper [49] presents the identification of apple leaf diseases with an accuracy of 93.71% using DenseNet-121. The study [16 ...
Mosaic, Rust, Brown spot, and Alternaria leaf spot are the four common types of apple leaf diseases. Early diagnosis and accurate identification of apple leaf diseases can control the spread of infection and ensure the healthy development of the apple industry. The existing research uses complex image preprocessing and cannot guarantee high recognition rates for apple leaf diseases. This paper ...
This research indicates that the proposed deep learning model provides a better solution in disease control for apple leaf diseases with high accuracy and a faster convergence rate, and that the image generation technique proposed in this paper can enhance the robustness of the convolutional neural network model. Expand.
The identification of apple leaf diseases is crucial to reduce yield reduction and timely take disease control measures. Employing deep learning for apple leaf disease identification is challenging because of the limited availability of samples for supervised training and the serious class imbalance. Hence, this paper proposes an accurate deep learning-based pipeline to solve the problem of ...
However, the existing research lacks an accurate and fast detector of apple diseases for ensuring the healthy development of the apple industry. This paper proposes a deep learning approach that is based on improved convolutional neural networks (CNNs) for the real-time detection of apple leaf diseases. In this paper, the apple leaf disease ...
Early detection and accurate diagnosis of leaf diseases can be significantly helpful in controlling their spread and improving the yield and quality of production. However, it is challenging to design and develop novel models to analyze complex, noise-contaminated leaf images and infer, preferably in real time, with high accuracy and precision. A Deep Convolutional Neural Network (DCNN) model ...
Early diagnosis and accurate identification of apple tree leaf diseases (ATLDs) can control the spread of infection, to reduce the use of chemical fertilizers and pesticides, improve the yield and quality of apple, and maintain the healthy development of apple cultivars. In order to improve the detection accuracy and efficiency, an early diagnosis method for ATLDs based on deep convolutional ...
The automatic detection of diseases in plants is necessary, as it reduces the tedious work of monitoring large farms and it will detect the disease at an early stage of its occurrence to minimize further degradation of plants. Besides the decline of plant health, a country's economy is highly affected by this scenario due to lower production. The current approach to identify diseases by an ...
Zhong Y Zhao M Research on deep learning in apple leaf disease recognition Comput Electron Agric 2020 168 10.1016/j.compag.2019.105146 Google Scholar Digital Library; Zhou C, Wu M, Lam SK (2019) SSA-CNN: semantic self-attention CNN for pedestrian detection, pp 4321-4330 Google Scholar
Second, we present a deep learning based real-time leaf disease detection system to identify seven types of diseases and pests that affect apple plants which can help fruit growers in accurately identifying various disease on time and provide helpful recommendations. 2. Dataset2.1. Apple leaf disease dataset
Therefore, it is of great significance to study the identification of apple leaf diseases. Based on DenseNet-121 deep convolution network, three methods of regression, multi-label classification and focus loss function were proposed to identify apple leaf diseases. In this paper, the apple leaf image data set, including 2462 images of six apple ...
Apple tree diseases have perplexed orchard farmers for several years. At present, numerous studies have investigated deep learning for fruit and vegetable crop disease detection. Because of the complexity and variety of apple leaf veins and the difficulty in judging similar diseases, a new target detection model of apple leaf diseases DF-Tiny-YOLO, based on deep learning, is proposed to ...
Apple leaf diseases threaten apple orchard sustain ability and production worldwide. Accurate and early identification is essential for the successful care and control of many disorders. This article suggests a convolutional neural network (CNN) model for classifying apple leaf diseases based on federated learning. Using a dataset of 7,832 photos with a resolution of 224×224 pixels and five ...
This paper proposes a deep learning approach that is based on improved convolutional neural networks (CNNs) for the real-time detection of apple leaf diseases. In this paper, the apple leaf disease dataset (ALDD), which is composed of laboratory images and complex images under real field conditions, is first constructed via data augmentation ...
Identify Apple Leaf Diseases Using Deep Learning Algorithm Figure 1. Part of images in the data set Figure 2. Images of disease symptoms on apple leave captured under different light conditions: indirect sunlight on the leaf (A), direct sunlight on the leaf (B), and strong reflection on the leaf (C). Figure 3. Sample images from the data
This research indicates that the proposed deep learning model provides a better solution in disease control for apple leaf diseases with high accuracy and a faster convergence rate, and that the image generation technique proposed in this paper can enhance the robustness of the convolutional neural network model.
Apple scab is spawned by the fungus Venturia inaequalis, and it has Olive-green and black velvety stain with misty margins on leaves. These are the types of diseases used in this study. The goal of this research is to identify diseases in frond by using Inception v3 one of the pre-trained models in CNN.
The proposed model aims in reducing complexity in classifying apple leaf disease using deep learning. The proposed system shows the best validation accuracy of 93.3% on the apple leaf disease dataset. This method outperforms some existing state-of-the-art. The processing time for each image is at an average of 14s.
Compared with previous studies done by scholars for apple leaf disease identification [29 ... etc. will also be the next step for effective implementation in subsequent research. With the deep learning technology in the field of target detection, several detection techniques have been effectively integrated, more and more new theories and ...
Researcher developed different deep learning techniques for crop disease identification, which worked on different dataset of crops. In this paper different rice plant diseases are described.
In this research, authors have developed plant disease recognition model to identify the disease in apple leaf and predicting the percentage of disease affected in leaf, based on leaf image classification and applying convolutional neural network algorithms. This model obtains an accuracy of 97.5%.
Food is the basic necessity for human survival and plant diseases act as a critical hindrance to the quality of harvested crops. Timely identification and administration of plant diseases are remarkably essential owing to the ever-increasing population and global climate change. Though many algorithms have been designed for early diagnosis of plant leaf diseases in existing literature, the ...
The specific kind of leaf disease that affects a plant's leaves is not always easy for the naked eye to discern. Therefore, cutting-edge technological methods including deep learning algorithms, machine learning, and artificial intelligence have been employed to improve the accuracy and recognition rate in order to safeguard plants from leaf ...